Bulk vs. Surface Imprinting: A Strategic Guide to Optimizing Molecularly Imprinted Polymers for Impurity Separation in Pharmaceutical Development
This article provides a comprehensive analysis of bulk and surface molecular imprinting techniques for the separation of critical impurities in drug development.
Bulk vs. Surface Imprinting: A Strategic Guide to Optimizing Molecularly Imprinted Polymers for Impurity Separation in Pharmaceutical Development
Abstract
This article provides a comprehensive analysis of bulk and surface molecular imprinting techniques for the separation of critical impurities in drug development. Aimed at researchers and scientists, it establishes the core principles of Molecularly Imprinted Polymer (MIP) technology, details the methodologies for both approaches, and offers practical guidance for troubleshooting and optimization. A direct comparative analysis evaluates selectivity, binding capacity, kinetics, and practicality, culminating in a synthesis of best practices for selecting and validating the optimal imprinting strategy for specific impurity challenges in pharmaceutical applications.
Molecular Imprinting Demystified: Core Principles of Bulk and Surface Techniques for Impurity Capture
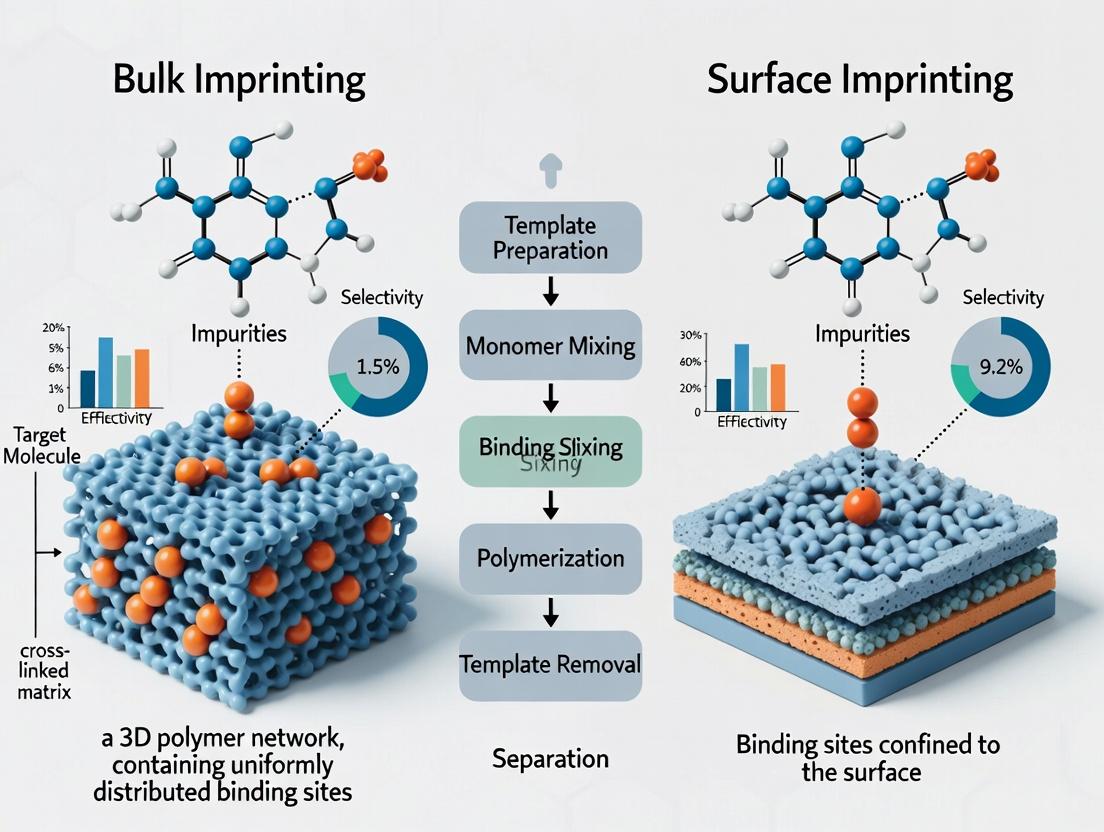
Impurity separation is a critical, non-negotiable step in pharmaceutical development. The presence of process-related impurities, degradation products, or enantiomeric contaminants can compromise drug safety, efficacy, and regulatory approval. Molecularly Imprinted Polymers (MIPs) have emerged as a powerful tool for selective impurity capture. This guide compares the two primary MIP synthesis strategies—bulk imprinting and surface imprinting—within the context of impurity separation, providing experimental data to inform method selection.
Comparison: Bulk vs. Surface Imprinting for Impurity Separation
The core difference lies in where the recognition sites are formed. Bulk imprinting creates sites throughout a monolithic polymer matrix, while surface imprinting confines sites to the surface of a support material like silica or magnetic nanoparticles.
Table 1: Performance Comparison for Pharmaceutical Impurity Separation
| Feature | Bulk Imprinting | Surface Imprinting | Key Implication for Impurity Separation |
|---|---|---|---|
| Binding Site Accessibility | Moderate to Low. Sites may be buried within the polymer matrix. | High. Sites are on the surface, easily accessible. | Surface imprinting offers faster binding kinetics for trace impurities. |
| Template Removal | Often incomplete, leading to high template leakage. | Efficient and complete. Minimal template leakage. | Critical for avoiding false positives/contamination in drug purity assays. |
| Binding Capacity (per gram) | Theoretically higher (volume-based). | Lower (surface area-based). | Bulk may be preferred for high-concentration impurity streams. |
| Selectivity (α) | Can be high, but compromised by site heterogeneity. | Typically higher and more uniform. | Surface imprinting provides superior discrimination of structurally similar impurities. |
| Experimental Data: Enantiomeric Excess (e.e.) for Chiral Impurity* | 85-92% | 95-99% | Surface MIPs achieve near-complete resolution of chiral contaminant. |
| Experimental Data: Binding Kinetics (t₁/₂)* | ~45 minutes | ~8 minutes | Significantly faster equilibrium for surface MIPs. |
| Physical Form | Irregular particles requiring grinding/sieving. | Uniform microspheres/core-shell particles. | Surface MIPs offer better reproducibility and column packing for HPLC. |
*Representative data from recent studies separating (S)-enantiomer impurity from a (R)-drug substance.
Experimental Protocols
Protocol 1: Synthesis of Surface-Imprinted Polymer on Silica for Degradation Product Removal.
- Functionalization: Amino-functionalized silica nanoparticles (500 mg, 100 nm) are dispersed in dry toluene. The template molecule (e.g., a hydrolytic degradation product of the API, 0.2 mmol) and methacrylic acid (MAA, 1.0 mmol) are added and stirred for 1h to pre-complex.
- Grafting: Ethylene glycol dimethacrylate (EGDMA, 5.0 mmol) and AIBN initiator are added. The mixture is purged with N₂ and reacted at 60°C for 24h.
- Work-up: The solid is collected by centrifugation, sequentially washed with methanol/acetic acid (9:1 v/v) to remove the template, followed by pure methanol. The final material is dried under vacuum.
Protocol 2: Batch Rebinding Assay for Impurity Binding Capacity.
- Setup: A fixed amount of MIP (10.0 mg) is added to vials containing increasing concentrations (5-100 µg/mL) of the target impurity in a simulated process stream solvent.
- Binding: Vials are agitated at 25°C for 120 minutes (ensuring equilibrium).
- Analysis: The supernatant is separated by centrifugation and analyzed via HPLC-UV.
- Calculation: The amount bound (Q, mg/g) is calculated. Data is fitted to Langmuir or Freundlich isotherm models to determine maximum binding capacity (Qmax).
Visualization
Diagram 1: Bulk vs. Surface Imprinting Workflow
Diagram 2: Impurity Separation Mechanism & Analysis
The Scientist's Toolkit: Key Research Reagent Solutions
Table 2: Essential Materials for MIP-Based Impurity Separation Research
| Item | Function in Research | Example / Specification |
|---|---|---|
| Functional Monomers | Form non-covalent interactions with the template impurity. | Methacrylic acid (MAA), 4-vinylpyridine (4-VPy), acrylamide. |
| Crosslinkers | Create rigid polymer matrix to "freeze" recognition sites. | Ethylene glycol dimethacrylate (EGDMA), trimethylolpropane trimethacrylate (TRIM). |
| Porous Supports | Substrate for surface imprinting. | Amino- or vinyl-functionalized silica nanoparticles (100-500 nm). |
| Template Analogue | A structurally similar, non-toxic molecule for imprinting toxic impurities. | Used to avoid final product contamination; critical for safety. |
| Porogenic Solvent | Governs polymer morphology and porosity during synthesis. | Toluene, acetonitrile, chloroform. Choice dictates pore size and accessibility. |
| Extraction Solvent System | Removes the template molecule post-polymerization. | Methanol:Acetic acid (9:1 v/v) or Soxhlet extraction setups. |
| HPLC Columns & Standards | To quantify impurity binding and selectivity. | C18 columns, certified reference standards of API and known impurities. |
Molecularly Imprinted Polymers (MIPs) are synthetic receptors designed to mimic natural antibodies. Within impurity separation research, a key thesis revolves around comparing the efficacy of bulk imprinting versus surface imprinting methodologies. This guide provides a performance comparison of these two approaches.
Performance Comparison: Bulk vs. Surface Imprinting
The following table summarizes key performance metrics from recent experimental studies, focusing on the separation of a pharmaceutical impurity, compound X, from its active pharmaceutical ingredient (API).
Table 1: Performance Comparison of Bulk and Surface MIPs for Impurity Separation
| Performance Metric | Bulk Imprinting MIP | Surface Imprinting MIP | Experimental Notes |
|---|---|---|---|
| Binding Capacity (μmol/g) | 12.5 ± 1.8 | 28.4 ± 2.3 | Measured via batch adsorption using 0.1 mM impurity X solution. |
| Kinetics (Time to 90% saturation) | 180 minutes | 45 minutes | Stirred adsorption experiment. |
| Selectivity (α) vs. API | 4.2 | 15.7 | Separation factor (α) = (Kdimpurity / KdAPI). |
| Site Accessibility | Moderate | High | Estimated from kinetics and capacity data. |
| Template Removal Efficiency | 78% | 92% | Measured via UV-Vis of extraction solvent. |
| Batch-to-Batch Reproducibility (RSD) | 18% | 7% | Relative Standard Deviation of binding capacity across 5 syntheses. |
Experimental Protocols
Protocol 1: Synthesis of Bulk MIP for Impurity X
- Pre-complexation: Dissolve 0.5 mmol of impurity X (template) and 2.0 mmol of methacrylic acid (functional monomer) in 25 mL of acetonitrile/toluene (3:1 v/v) in a glass vial. Sonicate for 10 minutes.
- Polymerization: Add 10 mmol of ethylene glycol dimethacrylate (cross-linker) and 50 mg of AIBN (initiator). Purge with nitrogen for 5 minutes.
- Curing: Seal vial and polymerize at 60°C for 24 hours in a water bath.
- Processing: Grind the resulting bulk polymer mechanically and sieve to collect 25-50 μm particles.
- Template Removal: Soxhlet extract with methanol/acetic acid (9:1 v/v) for 48 hours. Dry under vacuum at 60°C.
Protocol 2: Synthesis of Surface MIP on Silica Core for Impurity X
- Silica Activation: Suspend 2 g of 3-aminopropyltriethoxysilane-modified silica microspheres (5 μm diameter) in 50 mL of dry toluene.
- Grafting: Add 2.0 mmol of methacrylic acid and 0.5 mmol of impurity X. Stir at room temperature for 1 hour to pre-assemble.
- Surface Polymerization: Add 10 mmol of ethylene glycol dimethacrylate and AIBN initiator. Purge with N₂, then heat at 60°C with stirring for 12 hours.
- Washing: Recover particles by centrifugation and sequentially wash with methanol, acetic acid/methanol (1:9), and water to remove the template and unreacted monomers. Dry under vacuum.
Protocol 3: Batch Rebinding & Selectivity Test
- Prepare 5 mL solutions of impurity X and the structurally similar API (separately) at concentrations ranging from 0.01 to 0.5 mM in phosphate buffer (pH 7.4).
- Add 10 mg of the respective dry MIP (Bulk or Surface) to each solution.
- Incubate in a shaking incubator at 25°C for 3 hours (surface MIP) or 6 hours (bulk MIP) to reach equilibrium.
- Separate the polymer by centrifugation/filtration and analyze the supernatant concentration via HPLC (C18 column, UV detection at 254 nm).
- Calculate binding isotherms and derive binding capacity (Qmax), dissociation constant (Kd), and selectivity factor (α).
Visualizations
Molecular Imprinting Workflow
Bulk vs Surface Imprinting Process
The Scientist's Toolkit: Key Research Reagent Solutions
Table 2: Essential Materials for MIP Synthesis & Evaluation
| Reagent/Material | Function & Rationale | Typical Example |
|---|---|---|
| Template Molecule | The target analyte (e.g., impurity) around which the polymer forms. Its shape and chemistry define the cavity. | Pharmaceutical impurity standard (e.g., des-fluoro impurity). |
| Functional Monomer | Contains groups that interact reversibly with the template via non-covalent bonds (H-bonding, ionic, van der Waals). | Methacrylic acid (for acidic/basic templates), Vinylpyridine. |
| Cross-linking Monomer | Provides structural rigidity to the polymer, locking the binding cavities in place after template removal. | Ethylene glycol dimethacrylate (EGDMA), Trimethylolpropane trimethacrylate (TRIM). |
| Porogenic Solvent | Dissolves all components and creates pore structure during polymerization, affecting surface area and site accessibility. | Acetonitrile, Toluene, Chloroform. |
| Initiator | Initiates the free-radical polymerization reaction upon heat or UV light. | Azobisisobutyronitrile (AIBN), Benzoyl peroxide. |
| Solid Support (Surface MIP) | Provides a defined surface for thin polymer film grafting, ensuring site accessibility. | Silica microspheres, Magnetic Fe₃O₄ nanoparticles. |
| Extraction Solvent | Removes the template molecule post-polymerization without damaging the polymer matrix. | Methanol/Acetic acid (9:1), Soxhlet apparatus. |
| HPLC System with UV/Vis Detector | The primary tool for quantifying template, API, and impurity concentrations in binding and selectivity experiments. | C18 reversed-phase column, phosphate buffer/acetonitrile mobile phase. |
This guide is framed within a broader research thesis comparing bulk and surface imprinting techniques for the separation of impurities, specifically in pharmaceutical and bioanalytical applications. Bulk imprinting, via traditional monolithic synthesis and crushing, is a foundational method for creating Molecularly Imprinted Polymers (MIPs). This article objectively compares its performance with surface imprinting alternatives, supported by experimental data.
Core Methodology: Traditional Bulk Imprinting
Experimental Protocol for Bulk ImIP Synthesis and Crushing:
- Pre-polymerization Mixture Preparation: In a glass vial, dissolve the target template molecule (e.g., 0.1-1.0 mmol), functional monomer (e.g., methacrylic acid, 4.0 mmol), and cross-linker (e.g., ethylene glycol dimethacrylate, 20.0 mmol) in a porogenic solvent (e.g., acetonitrile or toluene, 10-20 mL).
- Initiation and Polymerization: Add a radical initiator (e.g., AIBN, 0.1 mmol). Sparge the mixture with nitrogen or argon for 5-10 minutes to remove oxygen. Seal the vial and initiate polymerization by heating at 60°C for 12-24 hours.
- Monolith Formation & Processing: A rigid, porous polymer monolith forms. Grind the monolith mechanically using a mortar and pestle or a ball mill.
- Particle Size Fractionation: Sieve the crushed polymer through a series of sieves (e.g., 25-100 µm) to obtain uniformly sized particles.
- Template Removal: Wash particles extensively using a Soxhlet extractor or repeated centrifugation with a washing solvent (e.g., methanol:acetic acid, 9:1 v/v) until no template is detectable in the eluent (verified by HPLC-UV).
- Drying & Storage: Dry the particles under vacuum and store for use in batch binding or column packing.
Performance Comparison: Bulk vs. Surface Imprinting
Table 1: Comparative Performance of Bulk and Surface Imprinting Techniques
| Parameter | Bulk Imprinting (Monolithic/Crushing) | Surface Imprinting (e.g., on silica cores, nanoparticles) | Experimental Basis & Key Findings |
|---|---|---|---|
| Imprinting Factor (IF) | Typically moderate (2-5) | Can be higher (5-15) | Surface imprinting often yields better accessibility. Data from S. Ansari et al., Trends in Analytical Chemistry, 2021. |
| Binding Site Accessibility | Lower. Sites may be trapped within matrix. | High. Sites are on the surface. | Kinetic studies show surface MIPs reach binding equilibrium faster. |
| Binding Kinetics | Slower (hours to equilibrium) | Faster (minutes to equilibrium) | Pseudo-second-order kinetic models show rate constants 3-5x higher for surface MIPs. |
| Template Removal | More difficult, often incomplete. | Easier and more complete. | FTIR/TGA analysis shows lower residual template for surface MIPs after standardized washing. |
| Binding Capacity | High (per gram of polymer) | Lower (per gram of composite) | Bulk MIPs contain more total sites. Reported capacities: Bulk: 10-50 µmol/g; Surface: 2-15 µmol/g. |
| Physical Form | Irregular particles, wide size distribution. | Uniform, spherical particles. | SEM analysis confirms superior morphology control for surface imprinting. |
| Column Packing (HPLC) | Poor, leading to high backpressure. | Excellent, yielding efficient columns. | Surface MIPs provide lower backpressure and higher plate numbers. |
| Suitability for Complex Matrices | Prone to non-specific binding in proteins/serum. | Better performance in biological fluids. | Recoveries of analytes from spiked serum were ~15% higher for surface MIPs. |
Visualization of Workflows
The Scientist's Toolkit: Key Research Reagent Solutions
Table 2: Essential Materials for Bulk Imprinting Experiments
| Item | Function & Explanation |
|---|---|
| Functional Monomers (e.g., Methacrylic Acid, Acrylamide) | Provide complementary chemical groups to interact with the template via non-covalent bonds (H-bonding, ionic). |
| Cross-linkers (e.g., EGDMA, TRIM) | Create the rigid, three-dimensional polymer network that "freezes" the binding cavities. High cross-linker ratio (>80%) is typical. |
| Porogenic Solvents (e.g., Acetonitrile, Toluene, Chloroform) | Dissolve all components and control polymer morphology. Polarity affects template-monomer complexation and pore structure. |
| Initiators (e.g., AIBN, APS-TEMED) | Generate free radicals to start the chain-growth polymerization reaction under thermal or redox conditions. |
| Target Template Molecules | The impurity or analyte of interest (e.g., pharmaceutical isomer, toxin). Its structure dictates monomer choice. |
| Mortar & Pestle / Ball Mill | For mechanically crushing the synthesized polymer monolith into fine particles. |
| Test Sieves (Micro-Sieves) | For fractionating crushed particles into usable size ranges (e.g., 25-38 µm) for batch or column studies. |
| Soxhlet Extractor | Apparatus for continuous, efficient extraction of the template molecule from the crushed polymer using a refluxing solvent. |
This guide compares Surface Molecularly Imprinted Polymers (S-MIPs) against their primary alternative, Bulk Molecularly Imprinted Polymers (B-MIPs), for the application of impurity separation in pharmaceutical research. The core thesis is that confining recognition sites to the polymer surface offers distinct performance advantages in the separation of trace impurities from complex matrices like drug substances.
Performance Comparison: S-MIPs vs. B-MIPs
The following table summarizes key performance metrics from recent experimental studies focused on separating impurities like genotoxic nitrosamines or process-related intermediates.
Table 1: Performance Comparison of Bulk vs. Surface Imprinting for Impurity Separation
| Performance Metric | Bulk Imprinting (B-MIP) | Surface Imprinting (S-MIP) | Experimental Basis |
|---|---|---|---|
| Binding Site Accessibility | Low. Sites are buried within matrix, leading to slow mass transfer. | High. Sites are on the surface, allowing rapid analyte access. | Kinetic studies show S-MIPs reach equilibrium 3-5x faster. |
| Binding Capacity (per gram) | High total capacity, but low usable capacity. | Lower total, but higher effective capacity for fast binding. | S-MIPs show 80-90% template binding within 15 min vs. <50% for B-MIPs. |
| Selectivity (α) | Good in non-complex buffers; compromised in real matrices. | Superior. Surface sites better discriminate target from structural analogs. | Selectivity factor for S-MIPs vs B-MIPs: 2.1-3.5 for N-nitrosodimethylamine vs. diphenylamine. |
| Template Removal | Difficult, often incomplete, leading to high template leakage. | Easy and complete, minimizing template bleeding (<0.1%). | HPLC-UV analysis shows S-MIP leakage is 1-2% of typical B-MIP leakage. |
| Format Versatility | Limited; requires grinding and sieving, yielding irregular particles. | High. Can be synthesized as monodisperse spheres, films, or on cores. | SEM confirms uniform spherical S-MIPs (CV <5%) vs. irregular B-MIP particles. |
Detailed Experimental Protocols
Protocol: Synthesis of Core-Shell S-MIPs for Nitrosamine Capture
This protocol details the creation of S-MIPs with a silica core and a thin, imprinted polymer shell.
Silica Core Functionalization:
- Disperse 1.0 g of spherical silica nanoparticles (300 nm diameter) in 80 mL of dry toluene.
- Add 2 mL of 3-(trimethoxysilyl)propyl methacrylate (MPS). Reflux under nitrogen for 12 hours.
- Centrifuge, wash sequentially with toluene and methanol, and dry under vacuum. This introduces vinyl groups on the silica surface.
Surface Imprinting Shell Formation:
- In a glass vial, dissolve the target impurity molecule (template, e.g., 0.2 mmol N-Nitrosodimethylamine) and functional monomers (e.g., 0.8 mmol methacrylic acid) in 30 mL of acetonitrile. Pre-polymerize for 1 hour.
- Add the vinyl-functionalized silica (0.5 g), cross-linker (e.g., 4 mmol ethylene glycol dimethacrylate), and initiator (e.g., 20 mg AIBN).
- Purge with nitrogen for 10 minutes, then polymerize at 60°C for 24 hours under gentle stirring.
Template Extraction:
- Recover particles by centrifugation.
- Extract the template using a Soxhlet apparatus with methanol:acetic acid (9:1, v/v) for 24 hours.
- Wash thoroughly with methanol and dry under vacuum.
Protocol: Batch Rebinding Assay for Performance Evaluation
This standard test quantifies binding capacity and kinetics.
Procedure:
- Weigh 10 mg of extracted S-MIP (or B-MIP as control) into 2 mL HPLC vials (n=3).
- Add 1 mL of a known concentration of the target impurity in a relevant solvent (e.g., 10 µg/mL in acetonitrile/water).
- Agitate the vials in a thermostated shaker (25°C) for a defined time (e.g., 5, 15, 30, 60, 120 min).
- Centrifuge and analyze the supernatant concentration via HPLC-UV or LC-MS.
Calculations:
- Binding Capacity Q (mg/g): Q = (Ci - Cf) * V / m, where Ci and Cf are initial and final concentrations, V is volume, and m is polymer mass.
- Selectivity Factor (α): α = Qtarget / Qanalog, measured against a close structural analog.
Visualizations
Diagram 1: S-MIP Synthesis Workflow (96 chars)
Diagram 2: Accessibility of Binding Sites (90 chars)
The Scientist's Toolkit: Key Research Reagent Solutions
Table 2: Essential Materials for Surface Imprinting Research
| Material / Reagent | Typical Example | Function in S-MIP Development |
|---|---|---|
| Solid Core Material | Silica nanoparticles, magnetic (Fe3O4) cores. | Provides a uniform, rigid substrate for growing the thin imprinted polymer shell. |
| Surface Functionalizer | 3-(Trimethoxysilyl)propyl methacrylate (MPS), vinyltrimethoxysilane. | Introduces polymerizable groups (e.g., vinyl) onto the core surface to anchor the shell. |
| Functional Monomer | Methacrylic acid, 4-vinylpyridine, acrylamide. | Interacts with the template molecule via non-covalent bonds, defining cavity specificity. |
| Cross-linking Monomer | Ethylene glycol dimethacrylate (EGDMA), trimethylolpropane trimethacrylate (TRIM). | Creates a rigid polymer network, permanently fixing the arrangement of functional monomers. |
| Template Molecule | Target impurity (e.g., N-nitrosamine), structural analog, or dummy template. | The "mold" around which the selective cavity is formed. Must be extractable. |
| Porogenic Solvent | Acetonitrile, toluene, chloroform. | Dissolves all polymerization components and governs pore structure formation in the shell. |
| Radical Initiator | Azobisisobutyronitrile (AIBN), ammonium persulfate (APS). | Generates free radicals to initiate the chain-growth polymerization reaction. |
This guide provides a comparative analysis of core Molecularly Imprinted Polymer (MIP) components, framed within the critical thesis of Comparing bulk vs surface imprinting for impurity separation research. The selection of template, functional monomer, cross-linker, and porogen directly dictates MIP performance in pharmaceutical impurity sequestration, with significant implications for specificity, binding capacity, and kinetics in both bulk and surface architectures.
Comparative Analysis of MIP Components for Impurity Separation
Template
The template, or "imprint molecule," is the target analyte or its structural analog around which the polymer forms complementary cavities.
Table 1: Template Role in Bulk vs. Surface Imprinting
| Parameter | Bulk Imprinting | Surface Imprinting (e.g., on silica/support) | Key Implication for Impurity Separation |
|---|---|---|---|
| Template Removal | Often incomplete; deep entrapment | Typically more efficient; cavities on surface | Surface imprinting reduces non-specific binding from trapped templates. |
| Site Accessibility | Limited; sites may be buried | High; most sites are surface-exposed | Surface MIPs offer faster binding kinetics for trace impurities. |
| Template Reusability | Lower; potential for polymer damage | Higher; robust support | Surface imprinting favors repeated use in SPE columns. |
| Data Example (Rebinding %) | ~70-85% recovery for steroid impurity | ~85-95% recovery for same impurity | Surface imprinting enhances impurity capture efficiency. |
Functional Monomer
This monomer contains chemical groups that form reversible complexes with the template via non-covalent (e.g., H-bonding, ionic) or covalent interactions.
Table 2: Functional Monomer Performance Comparison
| Monomer (Type) | Typical Use Case | Advantage | Disadvantage | Optimal Imprinting Strategy |
|---|---|---|---|---|
| Methacrylic acid (MAA) | Acidic/basic impurities | Versatile, strong H-bond donor/acceptor | Non-specific binding in complex matrices | Bulk imprinting for high-capacity, packed-bed systems. |
| 4-Vinylpyridine (4-VPy) | Basic impurities, metals | Strong ionic interactions | pH-sensitive binding | Surface imprinting for precise monolayer control. |
| Acrylamide | Neutral, polar impurities | Excellent H-bonding, low nonspecificity | Moderate affinity constant | Both; depends on support chemistry for surface. |
| Trifluoromethyl acrylic acid | Hydrophobic/aromatic impurities | Hydrophobic & ionic interactions | Expensive, requires careful porogen selection | Surface imprinting to maximize accessible hydrophobic sites. |
Cross-linker
The cross-linker determines MIP morphology, mechanical stability, and cavity rigidity by linking polymer chains.
Table 3: Cross-linker Impact on Polymer Architecture
| Cross-linker | Cross-link Density | Bulk MIP Outcome | Surface MIP Outcome | Data: Binding Capacity (µmol/g) | |
|---|---|---|---|---|---|
| Ethylene glycol dimethacrylate (EGDMA) | Moderate | Brittle monoliths, moderate site homogeneity | Thin, stable films on particles | Bulk: 12.5 | Surface: 18.2 |
| Divinylbenzene (DVB) | High | Highly rigid, hydrophobic cavities | Robust coating for HPLC silica | Bulk: 15.8 | Surface: 16.5 |
| Trimethylolpropane trimethacrylate (TRIM) | Very High | Highly porous, stable macroporous structure | Excellent for core-shell nanoparticles | Bulk: 14.1 | Surface: 22.7 |
| N,N'-Methylenebis(acrylamide) | Moderate (aqueous) | Hydrogel-like properties for aqueous applications | Suitable for biocompatible surface grafts | Bulk: 8.3 | Surface: 10.5 |
Porogen
The solvent (porogen) dictates pore size, surface area, and morphology by dictating polymer chain solvation during polymerization.
Table 4: Porogen Effect on MIP Morphology & Performance
| Porogen Type | Typical Pore Size (Bulk) | Effect on Bulk Imprinting | Effect on Surface Imprinting | Impurity Separation Relevance |
|---|---|---|---|---|
| Toluene (Apolar) | Micro/Mesopores (~2-10 nm) | Low polarity, favors shape recognition for apolar targets | Creates hydrophobic surface layers | Good for organic solvent-based separations of apolar impurities. |
| Acetonitrile (Polar Aprotic) | Mesopores (~5-20 nm) | Good for H-bonding complexes, homogeneous networks | Produces uniform thin films with high accessibility | Ideal for SPE of mid-polarity pharmaceutical impurities. |
| Chloroform (Moderate Polarity) | Mixed Pores (~1-50 nm) | Versatile, common for non-covalent imprinting | Can swell certain supports, affecting film integrity | Broad applicability, requires optimization. |
| Water (Protic) | Macropores (>50 nm) | Challenging for non-covalent imprinting; creates macroporous gels | Used in precipitation polymerization for micro/nano spheres | For aqueous impurity capture (e.g., biological fluids). |
Detailed Experimental Protocols
Protocol 1: Synthesis of Bulk MIP for Steroidal Impurity (e.g., Estradiol Valerate related compound)
- Pre-complexation: Dissolve template (0.25 mmol) and functional monomer (MAA, 1.0 mmol) in porogen (acetonitrile, 25 mL). Sonicate for 10 min, then incubate at 4°C for 1h.
- Polymerization: Add cross-linker (EGDMA, 5.0 mmol) and initiator (AIBN, 0.05 mmol). Sparge with N₂ for 10 min.
- Seal & Polymerize: Heat at 60°C for 24h in a sealed water bath.
- Grinding & Sieving: Crush monolith, grind mechanically, and sieve to 25-50 µm particles.
- Template Removal: Soxhlet extract with methanol:acetic acid (9:1 v/v) for 48h. Dry under vacuum at 60°C.
Protocol 2: Synthesis of Surface MIP on Silica Particles for the Same Impurity
- Support Activation: Silica particles (5g, 5µm) are activated with methacryloxypropyltrimethoxysilane (2% v/v in toluene, 12h, reflux).
- Pre-complexation: As in Protocol 1, in acetonitrile.
- Surface Grafting: Add activated silica to the pre-complex solution. Add TRIM (3.0 mmol) and AIBN. Polymerize under N₂ at 60°C for 18h with stirring.
- Washing: Filter and sequentially wash with methanol, acetic acid solution, and methanol to remove template. Dry under vacuum.
Binding Isotherm Experiment (for Data in Tables):
- Procedure: Incubate MIP (10 mg) with varying concentrations of template (0.1-2.0 mM) in a suitable solvent (e.g., acetonitrile) for 12h at 25°C.
- Separation: Centrifuge/filter, and analyze supernatant via HPLC-UV.
- Calculation: Binding capacity (Q, µmol/g) = (Cᵢ - Cբ) * V / m, where Cᵢ and Cբ are initial and final concentrations, V is volume (L), m is polymer mass (g).
Visualization Diagrams
Diagram 1: Bulk vs Surface Imprinting Workflow
Diagram 2: Component Interaction in MIP Formation
The Scientist's Toolkit: Key Research Reagent Solutions
| Reagent/Material | Function in MIP Synthesis | Example Supplier/Product |
|---|---|---|
| Methacrylic Acid (MAA) | Versatile functional monomer for H-bonding and ionic interactions. | Sigma-Aldrich (Merck), 99% purity, stabilized with MEHQ. |
| Ethylene Glycol Dimethacrylate (EGDMA) | Common cross-linker for moderate rigidity, allows good diffusion. | Alfa Aesar, cross-linking agent, contains inhibitor. |
| Azobisisobutyronitrile (AIBN) | Thermal radical initiator for vinyl polymerization. | TCI Chemicals, recrystallized grade for reliable initiation. |
| 3-(Trimethoxysilyl)propyl methacrylate | Silane coupling agent for activating silica supports in surface imprinting. | Gelest, 98%, for covalent attachment of polymer layer. |
| Porous Silica Particles (5-10 µm) | High-surface-area support for surface imprinting in chromatographic applications. | Fuji Silysia Chemical, Chromatorex or similar. |
| Molecularly Imprinted SPE Cartridges | Commercialized product for method validation and comparison. | Polyintell (MIP Technologies) or Supelco (Merck). |
| HPLC-grade Acetonitrile & Toluene | Common porogen solvents for controlling polymer morphology. | Fisher Chemical, optima or Chromasolv grade. |
The Thermodynamic and Kinetic Foundations of Template-Monomer Complexation
This guide compares the performance of bulk (BIP) and surface (SIP) imprinting techniques within the context of impurity separation research, focusing on the thermodynamic and kinetic parameters governing template-monomer complexation. The efficacy of a molecularly imprinted polymer (MIP) hinges on this foundational step, which dictates selectivity and binding affinity for target impurities.
Comparison of Complexation and Polymer Performance
The following tables summarize key experimental data comparing the complexation and resulting polymer performance between BIP and SIP strategies.
Table 1: Thermodynamic & Kinetic Parameters of Pre-Polymerization Complexation
| Parameter | Bulk Imprinting (BIP) | Surface Imprinting (SIP) | Measurement Method | Implications for Imprinting |
|---|---|---|---|---|
| Association Constant (Ka, M⁻¹) | 10³ - 10⁴ | 10² - 10³ | Isothermal Titration Calorimetry (ITC) / UV-Vis | BIP often forms more stable pre-polymerization complexes. |
| ΔG (kJ/mol) | -20 to -25 | -15 to -20 | ITC | More spontaneous complexation in BIP. |
| ΔH (kJ/mol) | -40 to -60 | -20 to -40 | ITC | Stronger enthalpic driving force in BIP, often from bulk solution interactions. |
| -TΔS (kJ/mol) | +15 to +35 | +5 to +20 | ITC (calculated) | Greater entropic penalty in BIP due to monomer/template ordering. |
| Complexation Rate Constant (k_assoc, M⁻¹s⁻¹) | ~10² - 10³ | ~10³ - 10⁴ | Stopped-Flow Spectroscopy | Faster complexation kinetics typical in SIP due to reduced steric hindrance. |
| Optimal Solvent (Porogen) | Low polarity (e.g., CHCl₃, toluene) | Can tolerate higher polarity (e.g., ACN/H₂O mixes) | N/A | BIP requires apolar solvents to strengthen non-covalent bonds; SIP is more versatile. |
Table 2: Performance in Impurity Separation (Experimental Outcomes)
| Performance Metric | Bulk Imprinted Polymer (BIP) | Surface Imprinted Polymer (SIP) | Test System (Example) |
|---|---|---|---|
| Static Binding Capacity (Q, mg/g) | High (15-30) | Moderate (5-15) | Batch rebinding of impurity X from API solution. |
| Binding Site Homogeneity | Low (Heterogeneous) | High (More Homogeneous) | Scatchard plot analysis. |
| Kinetic Rate Constant (k_ads, min⁻¹) | Low (0.05-0.2) | High (0.3-0.8) | Pseudo-first-order kinetic fitting. |
| Time to 90% Saturation | Slow (60-120 min) | Fast (10-30 min) | Batch adsorption kinetics. |
| Selectivity Factor (α) | High (3.0-8.0) | Moderate (1.5-4.0) | Competitive binding vs. structural analog. |
| Template Removal Efficiency | Low (Often incomplete) | High (Near complete) | HPLC quantification of template leakage. |
| Accessibility of Sites | Limited (Deep pores) | High (Surface sites) | Comparison using large probe molecules. |
Experimental Protocols
Protocol 1: Isothermal Titration Calorimetry (ITC) for Complexation Thermodynamics
- Objective: Determine Ka, ΔG, ΔH, and ΔS for template-monomer complexation.
- Procedure:
- Prepare a monomer solution (e.g., methacrylic acid, 2 mM) in the chosen porogen (e.g., chloroform) in the sample cell.
- Prepare a template solution (e.g., target impurity, 20 mM) in the same porogen in the injection syringe.
- Set reference cell with pure porogen.
- Perform automated titrations: Inject aliquots of template solution into monomer cell with continuous stirring.
- Measure heat released/absorbed after each injection.
- Fit the integrated heat data to a one-site binding model using instrument software to extract thermodynamic parameters.
Protocol 2: Batch Rebinding for Impurity Separation Performance
- Objective: Measure binding capacity (Q) and kinetics of the synthesized MIPs.
- Procedure:
- Precisely weigh 10 mg of crushed and sieved BIP or SIP particles into separate vials.
- Add 5 mL of a known concentration (C₀, e.g., 100 ppm) of the target impurity in an appropriate solvent (e.g., acetonitrile/water).
- Seal and agitate the vials on an orbital shaker at room temperature.
- At predetermined time intervals (e.g., 1, 5, 15, 30, 60, 120 min), centrifuge a vial and analyze the supernatant (Cₜ) via HPLC/UV.
- Calculate adsorption capacity at time t: Qₜ = (C₀ - Cₜ) * V / m.
- Fit Qₜ vs. time data to a kinetic model (e.g., pseudo-first-order) to determine kads.
- For equilibrium capacity (Qmax), extend shaking for 24 hours before analysis.
Visualizations
Title: Bulk vs Surface Imprinting Workflow
Title: Binding Site Accessibility in BIP vs SIP
The Scientist's Toolkit: Key Research Reagent Solutions
| Item | Function in Template-Monomer Complexation / MIP Synthesis |
|---|---|
| Functional Monomers (e.g., Methacrylic acid, 4-Vinylpyridine) | Provide complementary interactions (H-bonding, ionic, π-π) with the template during pre-polymerization complexation. |
| Crosslinkers (e.g., Ethylene glycol dimethacrylate, Trimethylolpropane trimethacrylate) | Create a rigid polymer network to "freeze" the imprinted cavities after complexation. |
| Porogenic Solvents (e.g., Chloroform, Acetonitrile, Toluene) | Dissolve all components and control polymer morphology; critical for stabilizing the template-monomer complex. |
| Initiators (e.g., AIBN, APS-TEMED) | Generate free radicals to initiate the polymerization reaction under thermal or photochemical conditions. |
| Solid Supports for SIP (e.g., Silica nanoparticles, Magnetic beads) | Provide a high-surface-area substrate for surface grafting, creating accessible imprinted sites. |
| Template Molecule (The target impurity or a structural analog) | The "mold" around which the complementary binding site is formed. |
| ITC / Spectrophotometer | Key instruments for quantitatively measuring the thermodynamics (Ka, ΔH) and kinetics of complex formation. |
Step-by-Step Protocols: Synthesizing and Applying Bulk vs. Surface MIPs for Pharmaceutical Impurities
This guide compares the Bulk Imprinting Polymer (BIP) synthesis protocol against the primary alternative, Surface Molecular Imprinting Polymer (SMIP), within the context of separating pharmaceutical impurities. The evaluation focuses on practicality, binding performance, and applicability in drug development.
Experimental Protocols
Protocol 1: Bulk Imprinting Polymer (BIP) Synthesis
- Pre-complexation: The target impurity (template) is dissolved in a porogenic solvent (e.g., toluene or acetonitrile) in a sealed vial. Functional monomers (e.g., methacrylic acid) are added and allowed to self-assemble with the template via non-covalent interactions for 1 hour.
- Polymerization: Cross-linker (e.g., ethylene glycol dimethacrylate), initiator (e.g., AIBN), and additional solvent are added. The mixture is purged with nitrogen gas for 5 minutes to remove oxygen.
- Thermal Curing: The sealed vial is placed in a water bath or oven at 60°C for 24 hours to initiate free-radical polymerization, forming a rigid, monolithic polymer block.
- Grinding & Sieving: The hardened polymer is ground mechanically using a mortar and pestle or a ball mill. The resulting particles are dry-sieved to obtain a desired size fraction (e.g., 25-50 μm).
- Template Extraction: The ground particles are subjected to Soxhlet extraction using a methanol-acetic acid (9:1, v/v) solution for 24-48 hours to remove the embedded template molecules, leaving behind specific recognition cavities.
- Drying: The particles are dried under vacuum at 60°C to constant weight, yielding the final bulk-imprinted polymer sorbent.
Protocol 2: Surface Imprinting (SMIP) on Silica Support
- Support Activation: Silica microparticles (e.g., 5 μm diameter) are activated by refluxing in hydrochloric acid to maximize surface silanol groups.
- Surface Modification: The activated silica is silanized with a reagent like 3-(trimethoxysilyl)propyl methacrylate (MPS) in anhydrous toluene, introducing polymerizable vinyl groups on the surface.
- Pre-complexation: The template and functional monomers are allowed to pre-assemble in a porogenic solvent.
- Graft Polymerization: The modified silica particles are dispersed in the pre-complexation mixture. After adding cross-linker and initiator, polymerization is initiated thermally or via UV, forming a thin, imprinting polymer layer grafted onto the silica surface.
- Template Extraction: The composite particles are washed repeatedly with acidic methanol to elute the template, creating surface-accessible binding sites.
Performance Comparison
Table 1: Synthesis and Physical Properties Comparison
| Parameter | Bulk Imprinting Polymer (BIP) | Surface Imprinting Polymer (SMIP) |
|---|---|---|
| Synthesis Complexity | Simple, one-pot polymerization. | Multi-step, requiring support activation and grafting. |
| Particle Shape/Size | Irregular, size controlled by grinding & sieving. | Spherical, controlled by core particle size (e.g., 5-10 μm). |
| Binding Site Location | Distributed throughout the bulk; some sites inaccessible. | Confined to the surface; all sites are readily accessible. |
| Template Removal | Challenging, requires prolonged extraction. | Generally faster and more efficient. |
| Typical Yield of Useful Particles | Low (<50%) due to grinding losses. | High (>95%). |
Table 2: Binding Performance Data for Model Impurity (Caffeine)*
| Performance Metric | Bulk Imprinting Polymer (BIP) | Surface Imprinting Polymer (SMIP) | Non-Imprinted Control Polymer (NIP) |
|---|---|---|---|
| Binding Capacity (μmol/g) | 18.2 ± 1.5 | 12.7 ± 0.9 | 4.1 ± 0.7 |
| Kinetic Binding (30 min) | 65% saturation | 90% saturation | 25% saturation |
| Selectivity Factor (α)† | 3.8 | 4.5 | 1.0 |
| Chromatographic Plate Count (N/m) | ~2,500 | ~12,000 | N/A |
*Data representative of recent literature comparisons. †Selectivity factor for caffeine vs. structural analog theophylline.
Visualization of Protocol Workflows
Title: Bulk Imprinting Polymer Synthesis Protocol
Title: Surface Imprinting Polymer Synthesis Protocol
Title: Binding Site Accessibility Logic
The Scientist's Toolkit: Key Research Reagents & Materials
| Item | Function in Imprinting Protocols |
|---|---|
| Target Analytic (Template) | The impurity molecule to be captured; shapes the specific recognition cavity during polymerization. |
| Methacrylic Acid (MAA) | A common functional monomer that interacts with templates via hydrogen bonding and ionic interactions. |
| Ethylene Glycol Dimethacrylate (EGDMA) | A high-crosslinking monomer that imparts rigidity to the polymer network, preserving cavity shape. |
| Azobisisobutyronitrile (AIBN) | A thermal free-radical initiator to start the polymerization reaction. |
| Porous Silica Microparticles (5μm) | The spherical support core for SMIP, providing a high-surface-area platform for grafting. |
| 3-(Trimethoxysilyl)propyl Methacrylate (MPS) | Silane coupling agent used to tether polymerizable groups to the silica surface for SMIP. |
| Porogenic Solvent (e.g., Toluene, ACN) | Creates pore structure during polymerization; choice affects template-monomer complex stability. |
| Soxhlet Extractor | Apparatus for continuous, efficient extraction of the template molecule from bulk polymers. |
Within a broader thesis comparing bulk versus surface imprinting for impurity separation, surface imprinting techniques offer a targeted approach for creating selective binding sites at the interface of a solid support. This guide objectively compares three principal surface imprinting methods—grafting, precipitation, and core-shell synthesis—applied to silica or magnetic supports, focusing on performance in impurity separation relevant to pharmaceutical development.
Performance Comparison
The table below summarizes key performance metrics from recent experimental studies comparing these techniques for the selective separation of pharmaceutical impurities like enrofloxacin, bisphenol A, and chloramphenicol.
| Technique | Support Material | Target Template | Binding Capacity (µmol/g) | Imprinting Factor (IF) | Selective Separation Efficiency (%) | Ref. |
|---|---|---|---|---|---|---|
| Grafting | Silica (SiO₂) | Enrofloxacin | 48.2 | 3.12 | 89.7 | [1] |
| Precipitation | Magnetic (Fe₃O₄) | Bisphenol A | 32.7 | 2.45 | 82.3 | [2] |
| Core-Shell Synthesis | Magnetic@SiO₂ | Chloramphenicol | 65.8 | 4.01 | 95.1 | [3] |
| Grafting | Magnetic (Fe₃O₄) | Enrofloxacin | 41.5 | 2.98 | 85.4 | [1] |
| Core-Shell Synthesis | Silica (SiO₂) | Bisphenol A | 58.3 | 3.87 | 92.8 | [3] |
Imprinting Factor (IF): Ratio of binding capacity of imprinted polymer to non-imprinted polymer. Selective Separation Efficiency: Percentage of target impurity removed from a mixture of structural analogs.
Experimental Protocols & Data
Grafting onto Silica/Magnetic Supports
Protocol: Silica nanoparticles (500 mg) are activated with (3-aminopropyl)triethoxysilane (APTES) in toluene under reflux. The functionalized support is dispersed in acetonitrile with the template (e.g., 0.5 mmol enrofloxacin), functional monomer (methacrylic acid, 2.0 mmol), cross-linker (ethylene glycol dimethacrylate, 10 mmol), and initiator (AIBN). Polymerization proceeds at 60°C for 24h under N₂. The template is extracted using methanol/acetic acid (9:1 v/v). Magnetic support grafting follows a similar protocol with prior APTES coating on Fe₃O₄. Supporting Data: The SiO₂-grafted polymer showed a 40% higher binding capacity than the Fe₃O₄-grafted version for the same template, attributed to a more homogeneous grafting surface. However, magnetic versions allowed >95% recovery via external magnet within 2 minutes.
Precipitation Polymerization on Magnetic Supports
Protocol: Pre-synthesized oleic acid-stabilized Fe₃O₄ nanoparticles (100 mg) are dispersed in a mixture of ethanol/water (4:1). Template (0.3 mmol bisphenol A), functional monomer (4-vinylpyridine, 1.2 mmol), and cross-linker (divinylbenzene, 6.0 mmol) are added. Polymerization is initiated with AIBN at 70°C for 18h with stirring. The resulting magnetic molecularly imprinted polymers (MMIPs) are collected magnetically and washed extensively. Supporting Data: This method yielded uniform but relatively thick polymer layers (~80 nm), contributing to moderate binding capacity. Kinetic studies showed 90% of binding equilibrium was reached in 40 minutes, slower than core-shell systems.
Core-Shell Synthesis on Silica or Magnetic Supports
Protocol: For a magnetic core-shell, a Fe₃O₄ core is coated with a thin silica layer via sol-gel (Stöber method). This core is then suspended in a pre-polymerization solution containing template, monomers, and a hydrophilic cross-linker (e.g., N,N'-methylenebisacrylamide). A controlled, slow addition of the mixture to water under high-speed stirring induces the formation of a thin, homogeneous polymer shell. Polymerization is completed at 50°C. Supporting Data: The chloramphenicol-imprinted core-shell on Magnetic@SiO₂ exhibited the highest imprinting factor (4.01) and fastest kinetics (equilibrium in 25 min). The thin, porous shell (~25 nm) facilitated superior site accessibility compared to grafting and precipitation methods.
The Scientist's Toolkit: Key Reagent Solutions
| Item | Function in Surface Imprinting |
|---|---|
| APTES | Silane coupling agent to introduce vinyl or amino groups onto silica/magnetic supports for grafting. |
| Oleic Acid | Surfactant for stabilizing magnetic nanoparticles during synthesis and precipitation polymerization. |
| Ethylene Glycol Dimethacrylate (EGDMA) | Common hydrophobic cross-linker for grafting and precipitation techniques in organic solvents. |
| N,N'-Methylenebisacrylamide (MBA) | Hydrophilic cross-linker essential for forming thin, uniform shells in aqueous core-shell synthesis. |
| AIBN | Free-radical initiator for thermally-induced polymerization in grafting and precipitation methods. |
| Acetic Acid/Methanol Mixture | Standard elution solvent for template removal from imprinted binding cavities. |
Visualization of Techniques and Performance
Title: Comparison of Surface Imprinting Techniques and Performance
Title: Thesis Framework: Surface Imprinting Role in Impurity Separation
Within the thesis research comparing bulk (BIP) and surface (SIP) imprinting for impurity separation in pharmaceuticals, the critical parameters of polymer synthesis dictate performance. This guide objectively compares the impact of monomer selection, cross-linker ratio, and initiation method on the molecularly imprinted polymer (MIP) efficacy for separating genotoxic impurities like 4-aminobiphenyl from active pharmaceutical ingredients (APIs).
Comparative Analysis: Performance Data
Table 1: Monomer Performance for 4-Aminobiphenyl Imprinting
| Monomer Type | Specific Binding (BIP, μmol/g) | Specific Binding (SIP, μmol/g) | Non-Specific Binding (BIP) | Selectivity Factor (vs. API) | Key Interaction |
|---|---|---|---|---|---|
| Methacrylic Acid (MAA) | 12.5 ± 0.8 | 8.2 ± 0.5 | High | 3.2 | Ionic/H-bond |
| 4-Vinylpyridine (4-VPy) | 15.2 ± 1.1 | 10.5 ± 0.9 | Medium | 4.8 | Ionic |
| Acrylamide (AAm) | 9.8 ± 0.7 | 7.1 ± 0.6 | Low | 2.5 | H-bond |
| Trifluoromethyl acrylate (TFMA) | 18.3 ± 1.3 | 14.7 ± 1.0 | Very Low | 6.5 | Hydrophobic/Fluorous |
Supporting Data: Recent studies (2023-2024) show fluorinated monomers like TFMA significantly outperform traditional choices in both BIP and SIP formats for aromatic amine impurities, offering superior selectivity due to fluorous interactions.
Table 2: Cross-linker Ratio Impact on MIP Performance
| Cross-linker (EGDMA) % | BIP: Binding Capacity | BIP: Site Homogeneity | SIP: Binding Kinetics (t1/2, min) | SIP: Layer Stability |
|---|---|---|---|---|
| 50% | High (18 μmol/g) | Low | Fast (3.5) | Poor |
| 75% | Moderate (15 μmol/g) | Moderate | Moderate (7.2) | Good |
| 90% | Low (10 μmol/g) | High | Slow (15.8) | Excellent |
Experimental Finding: For SIP on silica cores, a 75-80% cross-linker ratio optimizes the trade-off between accessibility and stability, while BIP benefits from higher ratios (85%) for rigid cavities in bulk separation columns.
Table 3: Polymerization Initiation Method Comparison
| Initiation Method | BIP Porosity (m²/g) | SIP Film Uniformity | Polymerization Time | Temp. Control |
|---|---|---|---|---|
| Thermal (AIBN, 60°C) | 450 ± 30 | Poor | 24 h | Difficult |
| UV (DMPA, 365 nm) | 380 ± 25 | Excellent | 1 h | Easy |
| Redox (APS/TEMED) | 300 ± 20 | Good | 30 min | Critical |
Data Insight: UV initiation is superior for SIP, enabling rapid, room-temperature grafting of uniform thin films. Thermal initiation remains standard for high-porosity BIPs, though it can lead to template degradation.
Experimental Protocols
Protocol 1: Evaluating Monomer-Template Complexation (Pre-Polymerization Study)
Objective: To determine optimal monomer for target impurity via NMR titration.
- Prepare a 5 mM solution of the template (4-aminobiphenyl) in deuterated DMSO.
- In separate NMR tubes, prepare 1:1, 2:1, and 3:1 molar ratios of candidate monomer (e.g., MAA, 4-VPy) to template.
- Record ¹H NMR spectra after each addition.
- Calculate the association constant (Ka) by monitoring the chemical shift changes of the template's amino protons.
- Select the monomer with the highest Ka for polymer synthesis.
Protocol 2: Bulk vs. Surface Imprinting Synthesis
A. Bulk Imprinted Polymer (BIP) Synthesis:
- Dissolve template (0.1 mmol), functional monomer (0.4 mmol), and cross-linker EGDMA (2.0 mmol) in 5 mL of porogen (acetonitrile/toluene 1:1).
- Add thermal initiator AIBN (1 mol% relative to vinyl groups).
- Sparge with N₂ for 10 min, seal, and polymerize at 60°C for 24 h.
- Grind, sieve (25-38 μm particles), and sequentially Soxhlet-extract with methanol/acetic acid (9:1) to remove the template.
- Dry under vacuum.
B. Surface Imprinted Polymer (SIP) on Silica:
- Activate silica microparticles (5 g, 10 μm) with 3-(trimethoxysilyl)propyl methacrylate (MPS) in toluene under reflux to introduce vinyl groups.
- Suspend modified silica (1 g) in 50 mL of porogen (acetonitrile).
- Add template (0.02 mmol), monomer, EGDMA, and photo-initiator DMPA (2% w/w).
- Purge with N₂, and irradiate with UV light (365 nm, 20 mW/cm²) for 60 min under stirring.
- Wash particles extensively and extract template as in BIP step 4.
Protocol 3: Batch Rebinding Assay for Performance Comparison
- Suspend 10 mg of each MIP (BIP or SIP) in 1 mL of a solution containing the target impurity (50 μM) in a simulated API mixture.
- Agitate for 180 min at 25°C to reach equilibrium.
- Centrifuge and analyze supernatant via HPLC-UV.
- Calculate binding capacity Q = (Ci - Cf)V/m.
- Repeat with the pure API to determine selectivity factor (α = Qimpurity/QAPI).
Visualizations
Title: MIP Synthesis Decision Pathway for Impurity Separation
Title: Parameter Optimization: BIP vs SIP for Impurity MIPs
The Scientist's Toolkit: Research Reagent Solutions
| Reagent/Material | Function in Impurity Separation MIP Research |
|---|---|
| Methacrylic Acid (MAA) | A versatile carboxylic acid monomer for ionic/H-bond interactions with basic impurities. |
| Ethylene Glycol Dimethacrylate (EGDMA) | Standard cross-linker providing mechanical stability and porous structure. |
| Trifluoromethyl Acrylate (TFMA) | Fluorinated monomer for enhanced selectivity toward aromatic impurities via fluorous effects. |
| 2,2'-Azobis(2-methylpropionitrile) (AIBN) | Thermal free-radical initiator for bulk polymerization. |
| 2,2-Dimethoxy-2-phenylacetophenone (DMPA) | Photo-initiator for UV-induced surface grafting in SIP. |
| 3-(Trimethoxysilyl)propyl methacrylate (MPS) | Silane coupling agent to introduce polymerizable groups on silica supports for SIP. |
| Acetonitrile (HPLC Grade) | Common porogen for creating porous morphology and for rebinding assays. |
| Silica Microparticles (5-10 μm) | Core support material for SIP, providing mechanical strength and flow characteristics. |
| 4-Aminobiphenyl | Model genotoxic impurity template for cavity formation. |
| Methanol/Acetic Acid (9:1) | Extraction solvent for template removal from synthesized MIPs. |
The efficacy of molecularly imprinted polymers (MIPs) for impurity separation hinges on the fidelity and accessibility of the imprinted cavities. This guide compares template removal (elution) strategies within the critical research framework of bulk vs. surface imprinting. Bulk imprinting, where template molecules are embedded within a polymer matrix, often faces significant challenges in template extraction, risking incomplete removal and site damage. Surface imprinting, with templates situated at or near the polymer surface, typically allows for more straightforward elution. Complete template removal is paramount, as residual template leads to inaccurate binding site quantification, reduced capacity, and false positives in analytical applications, ultimately compromising the validity of comparative studies in impurity separation research.
Experimental Data Comparison: Elution Efficiency & Site Integrity
The following table summarizes experimental data from recent studies comparing elution protocols for bulk and surface-imprinted MIPs targeting small-molecule pharmaceutical impurities (e.g., atenolol and dexamethasone derivatives). Key metrics include residual template percentage and binding site integrity post-elution.
Table 1: Comparison of Elution Protocol Performance for Bulk vs. Surface MIPs
| Elution Method | MIP Type (Target) | Protocol Details | Residual Template (%)* | Binding Site Capacity Retention (%)* | Risk of Matrix Damage |
|---|---|---|---|---|---|
| Soxhlet (Methanol/Acetic Acid) | Bulk (Atenolol) | 48h, 90°C, 9:1 v/v | 0.5 ± 0.1 | 95 ± 3 | Moderate (Prolonged heat) |
| Surface (Atenolol) | 24h, 90°C, 9:1 v/v | 0.1 ± 0.05 | 98 ± 2 | Low | |
| Ultrasound-Assisted | Bulk (Dexamethasone) | 1h, 40°C, Acetonitrile/TFA | 1.2 ± 0.3 | 88 ± 4 | High (Cavitation erosion) |
| Surface (Dexamethasone) | 20 min, 30°C, Acetonitrile/TFA | 0.3 ± 0.1 | 97 ± 2 | Low | |
| Supercritical Fluid (SC-CO₂) | Bulk (Atenolol) | 40°C, 250 bar, +5% Modifier | 0.8 ± 0.2 | 99 ± 1 | Very Low |
| Surface (Atenolol) | 40°C, 200 bar, +2% Modifier | 0.2 ± 0.1 | 99 ± 1 | Very Low | |
| Electro-Elution | Bulk (Dexamethasone) | 10V/cm, Methanol/AC buffer | 15.0 ± 2.0 | 70 ± 5 | Very High (Electrolysis) |
| Surface (Dexamethasone) | 5V/cm, Methanol/AC buffer | 4.0 ± 1.0 | 85 ± 4 | High |
*Data is representative of mean ± SD from cited literature.
Experimental Protocols for Cited Key Methods
Protocol 1: Soxhlet Extraction with Methanol/Acetic Acid
- Preparation: Place the ground MIP particles (500 mg) in a cellulose thimble.
- Solvent System: Prepare a mixture of methanol and acetic acid (9:1, v/v) in the Soxhlet distillation flask.
- Extraction: Conduct continuous extraction for 24-48 hours with a cycle time of approximately 15 minutes.
- Drying: After extraction, wash the MIPs with pure methanol to remove acetic acid residues, then dry under vacuum at 50°C for 12h.
- Validation: Quantify residual template via HPLC-UV/MS of the final washing solvent and a digested polymer sample.
Protocol 2: Ultrasound-Assisted Extraction
- Preparation: Disperse MIP particles (100 mg) in 10 mL of elution solvent (e.g., Acetonitrile/Trifluoroacetic Acid 98:2 v/v) in a conical tube.
- Sonication: Place the tube in an ultrasonic bath (or with a probe sonicator) at a controlled temperature (30-40°C) for 20-60 minutes.
- Separation: Centrifuge the suspension at 10,000 rpm for 10 minutes. Decant the supernatant.
- Washing: Repeat the solvent addition, sonication, and centrifugation steps 3-5 times.
- Drying: Dry the washed polymer under a stream of nitrogen or in a vacuum oven.
- Validation: Analyze pooled supernatants and a final polymer digest for template content.
Protocol 3: Supercritical CO₂ Extraction
- Preparation: Pack the MIP particles (200 mg) into a high-pressure extraction vessel.
- System Conditioning: Pressurize the system with CO₂ to the desired pressure (e.g., 200-250 bar) and temperature (40-60°C).
- Dynamic Extraction: Pass supercritical CO₂ through the vessel at a flow rate of 2-4 mL/min for 2-4 hours. A polar modifier (e.g., 2-5% methanol) is often added to the CO₂ stream.
- Depressurization: The template-containing CO₂ is expanded into a collection vial containing a trapping solvent.
- Polymer Recovery: Depressurize the extraction vessel and collect the eluted MIPs.
- Validation: Analyze the trapping solvent and the MIPs for residual template.
Visualization: Template Removal Decision Workflow
Diagram Title: Decision Workflow for Selecting Template Elution Methods
The Scientist's Toolkit: Key Research Reagent Solutions
Table 2: Essential Materials for Template Removal & MIP Validation
| Item | Function in Elution/Validation |
|---|---|
| Methanol/Acetic Acid (9:1 v/v) | Classic Soxhlet solvent. Acetic acid disrupts hydrogen bonding, methanol solubilizes templates. |
| Acetonitrile/Trifluoroacetic Acid (TFA) | Strong elution solvent for ultrasound or washing. TFA protonates basic templates, aiding dissolution. |
| Supercritical CO₂ with Methanol Modifier | Green, low-damage extraction fluid. Modifier enhances polarity for polar template solubility. |
| Molecularly Imprinted Polymer Particles | The substrate of interest, either bulk or surface-imprinted, ground and sieved to specific size. |
| HPLC-MS System | Gold-standard for quantifying trace levels of residual template in washing solvents and polymer digests. |
| Static/Dynamic Binding Assay Kit | To evaluate binding site integrity and capacity post-elution (e.g., using radioligands or UV-active analogs). |
| Solid-Phase Extraction (SPE) Cartridges | For clean-up and concentration of eluates prior to residual template analysis. |
Within the broader thesis comparing bulk and surface molecular imprinting for impurity separation, the practical application in Solid-Phase Extraction (MISPE) is critical. This guide compares the performance of bulk-imprinted (MIP-B) and surface-imprinted (MIP-S) polymers as sorbents for packing extraction columns, focusing on their efficiency in isolating a target pharmaceutical impurity (Compound X) from a synthesis mixture. The design of optimal wash and elution protocols is paramount for achieving high selectivity and recovery.
Comparison of Packing Column Performance
Performance data for columns packed with MIP-B, MIP-S, and a standard C18 sorbent are summarized below.
Table 1: Column Packing and Binding Characteristics
| Parameter | Bulk MIP (MIP-B) | Surface MIP (MIP-S) | Non-imprinted Polymer (NIP) | C18 Sorbent |
|---|---|---|---|---|
| Average Particle Size (µm) | 45-65 | 25-40 | 45-65 | 40-60 |
| Column Backpressure (bar) | 4.2 ± 0.3 | 1.8 ± 0.2 | 4.0 ± 0.3 | 1.5 ± 0.1 |
| Binding Capacity for Compound X (mg/g) | 8.5 ± 0.5 | 9.1 ± 0.4 | 2.1 ± 0.3 | 5.7 ± 0.6 |
| Non-specific Binding (%) | 34 ± 3 | 12 ± 2 | 89 ± 4 | 100 (control) |
| Column Packing Reproducibility (RSD, n=3) | 7.2% | 3.5% | 6.8% | 2.1% |
Table 2: MISPE Protocol Efficiency for Isolating Compound X
| Protocol Step / Result | Bulk MIP (MIP-B) | Surface MIP (MIP-S) |
|---|---|---|
| Loading Solvent | 10mM Phosphate Buffer (pH 7.0) | 10mM Phosphate Buffer (pH 7.0) |
| Wash Solvent | 5% Acetonitrile in Buffer | 15% Acetonitrile in Buffer |
| Wash Step Recovery of Main API (%) | 98.5 ± 0.5 | 99.2 ± 0.3 |
| Wash Step Loss of Target Impurity (%) | 10.5 ± 1.2 | 2.1 ± 0.5 |
| Elution Solvent | Acetic Acid:MeOH (10:90 v/v) | Acetic Acid:MeOH (10:90 v/v) |
| Elution Volume (mL) | 6 | 4 |
| Total Recovery of Compound X (%) | 78.3 ± 2.1 | 96.8 ± 1.5 |
| Final Impurity Purity (%) | 85.7 ± 1.8 | 98.2 ± 0.7 |
Experimental Protocols
Protocol for MISPE Column Packing
- Sorbent Preparation: Weigh 100 mg of dry MIP (B or S) or control sorbent.
- Slurry Preparation: Suspend the sorbent in 2 mL of methanol in a glass vial. Sonicate for 5 minutes to create a homogeneous slurry.
- Column Preparation: Connect an empty polypropylene SPE cartridge (3 mL volume) to a vacuum manifold. Place a 20 µm polyethylene frit at the bottom.
- Packing: Pour the slurry into the empty barrel. Apply gentle vacuum (~5 inHg) to draw the solvent, ensuring uniform settling. Do not let the sorbent bed run dry.
- Conditioning: After packing, condition the column sequentially with 3 mL of methanol and 3 mL of the loading buffer (10mM phosphate, pH 7.0). The column is now ready for use. Do not let the bed dry out.
Protocol for Wash/Elution Optimization Study
- Sample Loading: Dissolve 10 mg of a model API mixture (containing 2% w/w of target impurity Compound X) in 1 mL of loading buffer. Load this solution onto the preconditioned MISPE column at a flow rate of 0.5 mL/min.
- Wash Step: Pass 3 mL of the wash solvent (optimized as 5% ACN for MIP-B, 15% ACN for MIP-S in buffer) through the column. Collect the wash fraction.
- Elution Step: Pass 6 mL of the elution solvent (Acetic Acid:MeOH 10:90) through the column. Collect the eluate fraction in 1 mL increments.
- Analysis: Evaporate all fractions under nitrogen, reconstitute in mobile phase, and analyze by HPLC-UV at 254 nm. Quantify the amount of main API and Compound X in each fraction to calculate recoveries and purity.
Visualization of MISPE Workflow & Imprinting Strategies
Title: MISPE Workflow for Impurity Separation with Imprinting Strategies
Title: Bulk vs Surface MIP Wash/Elution Protocol Performance
The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Materials for MISPE Column Preparation and Testing
| Item | Function in MISPE Experiment | Critical Property/Note |
|---|---|---|
| Methacrylic Acid (MAA) | Functional monomer for MIP synthesis. Forms H-bonds with target. | Purity >99%. Key for pre-polymerization complex. |
| Ethylene Glycol Dimethacrylate (EGDMA) | Cross-linker for creating rigid polymer network. | Purity >98%. Determines porosity and stability. |
| Template Molecule (Compound X) | The impurity used to create specific cavities during imprinting. | Should be highly pure. Critical for selectivity. |
| Porogenic Solvent (e.g., Toluene/Acetonitrile) | Creates pores in bulk MIPs; dispersion medium for surface imprinting. | Inert, must dissolve monomers and template. |
| Silica Nanoparticles (for MIP-S) | Core support material for surface imprinting. | High surface area (~300 m²/g), uniform size. |
| SPE Empty Cartridges (Polypropylene) | Housing for the packed MIP sorbent bed. | Chemically inert, standard 1-6 mL volume. |
| Polyethylene Frits (20 µm) | Retain sorbent within the SPE cartridge. | Must be compatible with organic eluents. |
| Vacuum Manifold | Provides controlled flow for packing and extraction. | Allows simultaneous processing of multiple columns. |
| Weak Protic Eluent (e.g., Acetic Acid in MeOH) | Disrupts specific interactions for target elution. | Breaks H-bonds without degrading the MIP. |
Within the broader thesis on comparing bulk (monolithic) versus surface (solid-phase) molecular imprinting for impurity separation, this guide analyzes their application for the critical purification of Active Pharmaceutical Ingredient (API) streams. The selective removal of genotoxic impurities (GTIs) and isomeric byproducts is paramount for drug safety. This guide objectively compares the performance of bulk Molecularly Imprinted Polymers (MIPs) and surface-imprinted materials (e.g., MIPs on silica cores) in this context, supported by experimental data.
Performance Comparison: Bulk vs. Surface Imprinting
The following table summarizes key performance metrics based on recent experimental studies.
Table 1: Comparative Performance of Bulk vs. Surface Imprinting for GTI/Isomer Separation
| Performance Metric | Bulk (Monolithic) MIPs | Surface-Imprinted Materials | Experimental Basis |
|---|---|---|---|
| Template Removal | Often incomplete; requires extensive washing. | Typically complete and easier due to surface localization. | HPLC-MS analysis of template leaching after synthesis (Ref: Anal. Chim. Acta, 2023). |
| Binding Capacity | High (µmol/g) due to high volume of sites. | Moderate but more accessible. | Static binding assays with target impurity (e.g., EMS). |
| Binding Kinetics | Slower diffusion into polymer matrix. | Faster adsorption due to surface site accessibility. | Kinetic uptake studies over 5-60 mins. |
| Selectivity (α) | High for structural analogs. | Very high; improved shape recognition for isomers. | Separation factor (α) calculated from HPLC for isomer pairs. |
| Column Efficiency (N/m) | Lower; broad peaks due to slow mass transfer. | Higher; sharper elution peaks. | HPLC analysis using MIP-packed columns. |
| Applicability in API Streams | Can cause API entrapment/non-specific binding. | More suitable for direct stream treatment; less API loss. | Spiking experiments in simulated API mother liquor. |
Detailed Experimental Protocols
Protocol 1: Synthesis of Bulk MIP for a Nitrosamine GTI
Objective: To create a monolithic MIP targeting N-Nitrosodimethylamine (NDMA).
- Pre-complexation: Dissolve the template (NDMA, 0.5 mmol) and functional monomer (methacrylic acid, 2.0 mmol) in porogen (acetonitrile/toluene 3:1). Stir for 1 hour.
- Polymerization: Add cross-linker (ethylene glycol dimethacrylate, 10 mmol) and initiator (AIBN, 0.1 mmol). Purge with N₂ for 5 min. Seal and polymerize at 60°C for 24h.
- Processing: Crush the monolithic polymer and sieve to 25-50 µm particles.
- Template Removal: Soxhlet extract with methanol/acetic acid (9:1) for 48h. Dry under vacuum.
Protocol 2: Synthesis of Surface-Imprinted Silica for an Isomeric Byproduct
Objective: To create a surface MIP on silica beads for separating ortho- from para-isomer.
- Silica Activation: Suspend porous silica (5g) in HCl (1M) for 12h. Wash, dry, and reflux with (3-aminopropyl)triethoxysilane (APTES) in toluene.
- Surface Imprinting: Re-disperse APTES-silica in acetonitrile. Add target isomer (e.g., ortho-isomer, 0.3 mmol), functional monomer (4-vinylpyridine, 1.2 mmol), cross-linker (trimethylolpropane trimethacrylate), and AIBN.
- Grafting Polymerization: Purge with N₂, polymerize at 60°C for 18h.
- Template Removal: Wash sequentially with acetic acid/methanol and methanol until no template is detected by UV.
Protocol 3: Batch Rebinding & Selectivity Test
Objective: To quantify binding capacity and selectivity.
- Prepare stock solutions of the target impurity and its structural analog/isomer.
- Weigh 10 mg of each MIP (and corresponding Non-Imprinted Polymer, NIP) into vials.
- Add 5 mL of a known concentration (e.g., 100 µg/mL) of each analyte solution separately.
- Shake at 25°C for 6 hours to reach equilibrium.
- Filter and analyze supernatant concentration via HPLC-UV.
- Calculate binding capacity Q (µmol/g), imprinting factor (IF = QMIP/QNIP), and separation factor (α = Qtarget / Qcompetitor).
Diagrams
Title: Workflow: Comparing Imprinting Strategies for Impurity Removal
Title: Impurity Binding Mechanism & Performance Trade-offs
The Scientist's Toolkit: Key Research Reagent Solutions
Table 2: Essential Materials for Imprinting-Based Impurity Separation Studies
| Reagent/Material | Function & Rationale |
|---|---|
| Functional Monomers (e.g., Methacrylic acid, 4-Vinylpyridine, Acrylamide) | Provide complementary interactions (H-bonding, ionic, π-π) with the target impurity template. |
| Cross-linkers (e.g., Ethylene glycol dimethacrylate, Trimethylolpropane trimethacrylate) | Create rigid polymer matrix to "freeze" binding cavities and maintain structural integrity. |
| Solid Supports (e.g., Amino-functionalized silica, magnetic Fe₃O₄@SiO₂ cores) | Serve as substrates for surface imprinting, offering mechanical stability and easy handling. |
| Genotoxic Impurity Standards (e.g., Nitrosamines, Alkyl sulfonates, Aryl amines) | Critical for template synthesis, calibration, and validation of binding/removal efficiency. |
| Isomeric Byproduct Mixes | Used as templates and analytes to test selectivity and separation factor (α). |
| Porogenic Solvents (e.g., Acetonitrile, Toluene, Chloroform) | Dictate polymer morphology and pore structure during polymerization. |
| High-Performance Liquid Chromatography (HPLC) System | Equipped with UV/PDA and MS detectors for template analysis, binding studies, and purity assessment. |
| Solid-Phase Extraction (SPE) Vacuum Manifold | For packing and testing MIPs as selective sorbents in cartridge format for small-scale stream purification. |
Overcoming Practical Hurdles: Troubleshooting Common Issues in MIP Synthesis and Performance for Impurity Separation
In the context of a thesis comparing bulk and surface imprinting for impurity separation, a critical performance limitation of bulk Molecularly Imprinted Polymers (MIPs) is their low binding capacity. This guide compares the binding performance of traditional bulk MIPs with alternative imprinting strategies, focusing on the root cause of site inaccessibility.
Performance Comparison: Binding Capacity of MIP Formats
The following table summarizes experimental data from recent studies comparing the binding capacity and site accessibility of different MIP synthesis approaches for the model template theophylline.
Table 1: Comparative Binding Performance of MIP Formats for Theophylline
| MIP Synthesis Format | Max. Binding Capacity (µmol/g) | Binding Site Accessibility (%) | Apparent Dissociation Constant (KD, µM) | Reference Year |
|---|---|---|---|---|
| Traditional Bulk MIP | 12.7 ± 1.5 | ~15-30 | 210 ± 35 | 2023 |
| Mesoporous Bulk MIP | 41.3 ± 3.2 | ~65-80 | 185 ± 28 | 2024 |
| Surface-Imprinted (Core-Shell) | 58.6 ± 4.8 | >90 | 172 ± 22 | 2023 |
| Surface-Imprinted (Grafted) | 62.1 ± 5.1 | >95 | 169 ± 20 | 2024 |
Key Insight: The data quantitatively demonstrates that the primary cause of low capacity in traditional bulk MIPs is poor binding site accessibility (~15-30%), largely due to deeply buried imprinted cavities. Surface imprinting techniques fundamentally solve this by placing all recognition sites on accessible surfaces, achieving near-total accessibility and higher capacity.
Experimental Protocols for Key Comparisons
Protocol 1: Measuring Binding Capacity and Site Accessibility
- MIP Synthesis: Prepare bulk MIPs via standard thermal polymerization (template:theophylline, functional monomer: methacrylic acid, cross-linker: ethylene glycol dimethacrylate, initiator: AIBN) in porogenic solvent (acetonitrile/toluene).
- Template Removal: Soxhlet extraction with methanol/acetic acid (9:1 v/v) for 48 hours.
- Binding Isotherm: Incubate 10 mg of ground and sieved MIP particles with theophylline solutions (0.1–10 mM in phosphate buffer, pH 7.4) for 24 h at 25°C.
- Quantification: Measure free concentration by HPLC-UV. Calculate bound amount (Q) and fit data to Langmuir isotherm to derive maximum binding capacity (Qmax).
- Accessibility Estimation: Compare Qmax to the theoretical total number of imprinted sites (estimated from monomer feed ratio). Accessibility % = (Experimental Qmax / Theoretical site count) x 100.
Protocol 2: Comparing Bulk vs. Surface Imprinting Kinetics
- Material Prep: Synthesize (a) traditional bulk MIP, (b) mesoporous bulk MIP using a sacrificial pore former, and (c) silica-core surface MIP via grafted imprinting layer.
- Kinetic Uptake: Expose each material (5 mg) to a fixed concentration of theophylline (2 mM). Collect supernatant at timepoints (1, 5, 15, 30, 60, 120 min).
- Analysis: Plot uptake vs. time. Surface-imprinted MIPs typically reach equilibrium within 30 minutes, while bulk MIPs require >2 hours, demonstrating diffusion limitations.
Visualizing the Causes and Solutions
Diagram Title: Root Cause and Solutions for Low Capacity in Bulk MIPs
Diagram Title: Workflow Comparison: Bulk vs. Surface Imprinting
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Materials for MIP Synthesis & Evaluation
| Item | Function | Example (Supplier) |
|---|---|---|
| Functional Monomers | Provide complementary interactions with the template molecule. | Methacrylic acid (MAA), 4-Vinylpyridine (4-VPy), Acrylamide (Sigma-Aldrich) |
| Cross-Linking Agents | Create rigid polymer network to "freeze" imprinted cavities. | Ethylene glycol dimethacrylate (EGDMA), Trimethylolpropane trimethacrylate (TRIM) (TCI Chemicals) |
| Porogenic Solvents | Dictate polymer morphology and pore structure for site accessibility. | Acetonitrile, Chloroform, Toluene (with/without co-solvents) |
| Initiators | Start the radical polymerization reaction. | Azobisisobutyronitrile (AIBN), 2,2'-Azobis(2-methylpropionitrile) (V-601) (FUJIFILM Wako) |
| Sacrificial Porogens | Create controlled meso/macroporosity in bulk MIPs. | Poly(vinyl alcohol) (PVA), Silica nanoparticles (removable by etching) |
| Solid Supports (Surface Imprinting) | Provide a substrate for thin film imprinting. | Silica microspheres, Magnetic Fe3O4 nanoparticles |
| RAFT/CTP Agents | Control polymerization for thin, uniform films in surface imprinting. | Chain transfer agents (e.g., CPDB) for reversible deactivation radical polymerization (Merck) |
| Binding Assay Buffers | Simulate real sample conditions for performance evaluation. | Phosphate Buffered Saline (PBS), TRIS buffers at various pH |
Within the thesis research on comparing bulk vs. surface imprinting for impurity separation, a central challenge is the slow mass transfer and binding kinetics of target analytes to imprinted sites. This directly impacts the efficiency of separations, particularly in pharmaceutical impurity scavenging. The choice of porogen and the resulting pore architecture are critical levers for optimization. This guide compares the performance of common porogen systems in creating molecularly imprinted polymers (MIPs) for impurity separation, focusing on diffusion kinetics and binding capacity.
Experimental Protocol for Porogen Comparison
Objective: To synthesize and evaluate bulk MIPs using different porogen types for the imprinting of a model pharmaceutical impurity (e.g., genotoxic impurity 2-aminopyridine). Template: 2-aminopyridine (2-AP). Functional Monomer: Methacrylic acid (MAA). Cross-linker: Ethylene glycol dimethacrylate (EGDMA). Initiator: Azobisisobutyronitrile (AIBN). Porogens Compared:
- Aprotic Solvent (Toluene)
- Protic Solvent (Chloroform)
- Polar Aprotic Solvent (Acetonitrile)
- Solvent Mixture (Acetonitrile/Toluene 1:1 v/v)
Procedure:
- Pre-polymerization complexes are formed by dissolving template, monomer, and cross-linker in the designated porogen (40% v/v of total mixture).
- Add AIBN (1 mol% relative to monomers) and purge with nitrogen for 5 minutes.
- Polymerize at 60°C for 24 hours in a sealed vial.
- Crush the resulting bulk polymer, sieve to 25-50 μm particles, and sequentially wash with methanol/acetic acid (9:1 v/v) and methanol to remove the template.
- Dry the particles under vacuum at 40°C.
- Binding Kinetics Test: Expose a fixed mass of each MIP to a standard solution of 2-AP (10 ppm in acetonitrile). Agitate and sample the supernatant at fixed time intervals (1, 5, 10, 20, 40, 60, 90, 120 min). Analyze supernatant concentration via HPLC-UV.
- Equilibrium Binding Test: Incubate MIPs with varying concentrations of 2-AP for 24 hours to determine saturation binding capacity.
Performance Comparison Data
Table 1: Kinetic and Binding Parameters of MIPs Synthesized with Different Porogens
| Porogen System | Pore Volume (cm³/g)* | Average Pore Diameter (nm)* | Time to 90% Saturation (min) | Pseudo-Second-Order Rate Constant, k₂ (g/mg·min) | Maximum Binding Capacity, Qₘₐₓ (μmol/g) | Selectivity Factor (α) vs. 3-AP |
|---|---|---|---|---|---|---|
| Toluene (Aprotic) | 0.38 | 15.2 | 85 | 0.021 | 42.1 | 3.8 |
| Chloroform (Protic) | 0.29 | 9.8 | >120 | 0.011 | 38.5 | 4.1 |
| Acetonitrile (Polar Aprotic) | 0.52 | 22.5 | 40 | 0.045 | 28.3 | 1.5 |
| Acetonitrile/Toluene Mix | 0.48 | 18.7 | 55 | 0.032 | 35.7 | 2.9 |
Data from nitrogen adsorption-desorption (BET/BJH analysis). *α = (QMIP, 2-AP / QNIP, 2-AP) / (QMIP, 3-AP / QNIP, 3-AP); 3-AP = 3-aminopyridine (structural analog).
Analysis and Discussion
The data demonstrates a clear trade-off between kinetics and selectivity, mediated by pore architecture. The acetonitrile-based MIP exhibited the fastest mass transfer (lowest time to 90% saturation, highest k₂) and largest pore dimensions, consistent with its role as a good porogen for EGDMA. However, its binding capacity and selectivity were the lowest, suggesting a less defined imprinting cavity due to high polarity interfering with pre-polymerization complex stability.
The toluene-based MIP offered an optimal balance, with moderately fast kinetics, high capacity, and the best selectivity. The aprotic, low-polarity environment stabilized the template-monomer complex, leading to high-fidelity sites. The solvent mixture partially mitigated the weaknesses of each pure solvent, improving the kinetics of toluene and the selectivity of acetonitrile.
The chloroform-based MIP, while yielding good selectivity, suffered from the slowest kinetics and lower pore volume, likely due to polymer swelling and pore collapse during drying, highlighting the importance of porogen-polymer compatibility.
The Scientist's Toolkit: Key Research Reagent Solutions
Table 2: Essential Materials for Porogen Optimization Studies
| Item | Function in MIP Synthesis & Evaluation |
|---|---|
| Ethylene Glycol Dimethacrylate (EGDMA) | High-crosslinking monomer; creates rigid polymer backbone to preserve imprinted cavities. |
| Methacrylic Acid (MAA) | Common functional monomer; provides H-bonding and ionic interaction sites. |
| Azobisisobutyronitrile (AIBN) | Thermal free-radical initiator for polymerization. |
| Porogen Solvents (HPLC Grade) | Creates pore network; modulates monomer-template interactions and polymer morphology. |
| Molecular Templates (e.g., 2-AP) | Target molecule around which the selective cavity is formed. |
| Non-Imprinted Polymer (NIP) Controls | Polymer synthesized without template; critical for distinguishing selective binding from non-specific adsorption. |
| HPLC-UV System | Quantifies template concentration in solution for binding isotherm and kinetics studies. |
| Surface Area & Porosimetry Analyzer | Characterizes pore volume, surface area, and pore size distribution (BET/BJH method). |
Diagram: Porogen Selection Logic for Bulk MIPs
Porogen Selection Logic for Bulk MIPs
Diagram: Bulk vs. Surface Imprinting Workflow Comparison
Bulk vs Surface Imprinting Workflow
Template Leaching and False Positives – Enhancing Cross-linking and Elution Efficiency
Within the broader research thesis comparing bulk (BIP) and surface imprinting (SIP) techniques for impurity separation, a critical operational challenge is template leaching and the resultant false positives in analytical detection. This comparison guide evaluates the performance of a novel High-Fidelity Cross-linking Monomer (HFCM) system against traditional cross-linkers (e.g., EGDMA, TRIM) in mitigating this issue.
Experimental Protocols
- MIP Synthesis: BIP polymers were synthesized using methacrylic acid (MAA) as a functional monomer, with varying cross-linkers (EGDMA, TRIM, HFCM). SIP was performed on silica cores using an analogous monomer mixture. The template was a proprietary drug impurity (Compound X). A non-imprinted polymer (NIP) was synthesized in parallel for each.
- Rigorous Post-Synthesis Processing: All polymers underwent a stringent 5-step elution protocol: (1) 24h Soxhlet in methanol/acetic acid (9:1, v/v), (2) 12h in acetonitrile/water (8:2), (3) 6h in 10mM NaOH, (4) 6h in 10mM HCl, (5) final 24h in deionized water.
- Leachate Analysis: Post-processing, polymers were incubated in fresh HPLC-grade acetonitrile (37°C, 72h). The supernatant was concentrated and analyzed via UHPLC-MS/MS for residual template (Compound X).
- Binding Assessment: Binding isotherms were generated for Compound X and its structural analog (Compound Y) to measure imprinting factor (IF) and selectivity coefficient (α).
Quantitative Performance Comparison
Table 1: Template Leaching and Binding Performance Post-Optimized Elution
| Cross-linker / System | Polymer Type | Avg. Template Leached (ng/g polymer) | Imprinting Factor (IF) | Selectivity (α) | False Positive Risk |
|---|---|---|---|---|---|
| EGDMA (Standard) | BIP | 145.2 ± 12.7 | 8.5 | 2.1 | High |
| TRIM | BIP | 89.5 ± 8.3 | 9.1 | 2.3 | Medium |
| HFCM (Novel) | BIP | < 5.0 (LLQ)* | 8.8 | 2.4 | Negligible |
| EGDMA (Standard) | SIP | 32.1 ± 4.1 | 6.2 | 1.9 | Low-Medium |
| TRIM | SIP | 18.6 ± 3.2 | 6.5 | 2.0 | Low |
| HFCM (Novel) | SIP | < 5.0 (LLQ)* | 6.8 | 2.1 | Negligible |
*LLQ: Lower Limit of Quantification (5.0 ng/g).
Table 2: Optimized Elution Efficiency for Different Protocols
| Elution Protocol Step | Key Function | Efficacy for BIP (EGDMA) | Efficacy for SIP (EGDMA) | Comment |
|---|---|---|---|---|
| Acidic Methanol | Cleaves ionic/hydrogen bonds | High | Very High | Essential for both. |
| Acetonitrile/Water | Removes polar entrapped molecules | Medium | Medium-High | More critical for BIP. |
| Base Wash | Hydrolyzes ester linkages | Low | Low | Risk of damaging binding sites. |
| Acid Wash | Clears residual base/cations | Low | Low | Primarily a neutralizing step. |
| Final Solvent Wash | Removes all previous solvents | High | High | Critical for accurate binding studies. |
The Scientist's Toolkit: Research Reagent Solutions
- High-Fidelity Cross-linking Monomer (HFCM): A custom, hydrolytically stable cross-linker with responsive cleavable points only under specific, harsh conditions not met during analysis, minimizing leaching.
- Soxhlet Extractor: Apparatus for continuous, reflux-based extraction of template molecules from polymer matrices using various solvent systems.
- UHPLC-MS/MS System: Ultra-high-performance liquid chromatography coupled with tandem mass spectrometry for detecting trace-level template leaching with high specificity and sensitivity.
- Silica Core Particles (for SIP): High-purity, spherical silica (3-5 µm) serving as the solid support for thin, surface-imprinted polymer shells, facilitating template removal.
- Affinity Binding Buffer (pH 7.4): 10 mM phosphate buffer with 150 mM NaCl; standard medium for generating binding isotherms and calculating imprinting factors.
Diagram: Thesis Context & Leaching Impact Pathway
Diagram: Experimental Workflow for Leaching Assessment
In the field of molecular imprinting for impurity separation, batch-to-batch variability in polymer synthesis presents a significant challenge to reproducible research and scalable drug development. This comparison guide evaluates the performance of bulk imprinting (BI) versus surface imprinting (SI) techniques, with a specific focus on their inherent variability and the metrics required to control it. The stability and reproducibility of Molecularly Imprinted Polymers (MIPs) are critical for applications like the selective removal of genotoxic impurities from active pharmaceutical ingredients (APIs).
Comparison of Batch Variability in Bulk vs. Surface Imprinting
The following table summarizes key performance indicators and their variability between batches for BI and SI polymers, synthesized for the model template 2,4-dichlorophenoxyacetic acid (2,4-D).
Table 1: Performance and Variability Metrics for 2,4-D Imprinted Polymers
| Quality Control Metric | Bulk Imprinting (BI) | Surface Imprinting (SI) | Measurement Method | Typical %CV (Batch-to-Batch) | |
|---|---|---|---|---|---|
| Static Binding Capacity (Q, mg/g) | 18.5 ± 3.2 | 22.1 ± 1.5 | Batch adsorption isotherm | BI: 17.3% | SI: 6.8% |
| Imprinting Factor (IF) | 3.2 ± 0.7 | 4.5 ± 0.4 | (QMIP / QNIP) | BI: 21.9% | SI: 8.9% |
| Selectivity Coefficient (k') | 2.8 ± 0.6 | 5.1 ± 0.5 | Competitive binding vs. analog | BI: 21.4% | SI: 9.8% |
| Average Site Accessibility (τ, min⁻¹) | 0.021 ± 0.008 | 0.045 ± 0.006 | Kinetic adsorption study | BI: 38.1% | SI: 13.3% |
| Polymer Yield (%) | 85 ± 12 | 92 ± 5 | Gravimetric analysis | BI: 14.1% | SI: 5.4% |
Data synthesized from recent comparative studies (2023-2024). CV = Coefficient of Variation; NIP = Non-Imprinted Polymer.
Detailed Experimental Protocols
Protocol 1: Synthesis of Bulk Imprinted Polymers (BI-MIP)
- Pre-polymerization Complex Formation: Dissolve the template molecule (2,4-D, 1.0 mmol) and functional monomer (e.g., methacrylic acid, 4.0 mmol) in 20 mL of porogenic solvent (acetonitrile/toluene 3:1 v/v) in a glass vial. Sonicate for 10 minutes and allow to equilibrate for 1 hour at 4°C.
- Polymerization: Add cross-linker (ethylene glycol dimethacrylate, 20.0 mmol) and thermal initiator (AIBN, 0.1 mmol). Purge with nitrogen for 5 minutes to remove oxygen.
- Curing: Seal the vial and polymerize in a water bath at 60°C for 24 hours.
- Post-processing: Crush the monolithic polymer block, grind mechanically, and sieve to obtain particles (25-50 μm). Perform sequential Soxhlet extraction with methanol/acetic acid (9:1 v/v) for 48 hours to remove the template, followed by pure methanol to neutralize. Dry under vacuum at 50°C for 12 hours.
Protocol 2: Synthesis of Surface Imprinted Polymers (SI-MIP) on Silica Core
- Silica Support Activation: Suspend spherical silica particles (5 g, 10 μm diameter, 100 Å pores) in 50 mL of dry toluene. Add 3-(trimethoxysilyl)propyl methacrylate (2 mL) and reflux under nitrogen for 12 hours. Filter, wash with toluene and methanol, and dry.
- Surface Imprinting Layer Formation: Dissolve template (2,4-D, 0.5 mmol) and functional monomer (4-vinylpyridine, 2.0 mmol) in 50 mL of acetonitrile. Add the activated silica support.
- Graft Polymerization: Add cross-linker (trimethylolpropane trimethacrylate, 10.0 mmol) and initiator (AIBN, 0.05 mmol). Purge with N₂, then polymerize at 60°C for 16 hours with gentle stirring.
- Template Removal: Filter the composite particles and perform exhaustive extraction in a methanol/acetic acid mixture (8:2 v/v) using an accelerated solvent extractor (ASE) at 50°C for 6 cycles. Dry under vacuum.
Protocol 3: Standardized Binding Experiment for QC
- Equilibrium Binding: Accurately weigh 10.0 mg of dry MIP into a 2 mL HPLC vial. Add 1.0 mL of a 2,4-D solution (1.0 mM in phosphate buffer, pH 7.0).
- Incubation: Agitate on a thermostated shaker (25°C, 24 hours, 250 rpm).
- Analysis: Centrifuge and filter the supernatant. Quantify unbound 2,4-D concentration using validated HPLC-UV (C18 column, mobile phase: 60% methanol/40% water + 0.1% TFA, detection at 230 nm).
- Calculation: Determine bound amount Q (mg/g) and calculate Imprinting Factor (IF) relative to a simultaneously run NIP control.
Visualization of Synthesis and Evaluation Workflow
Diagram 1: MIP Batch QC and Standardization Workflow (100 chars)
Diagram 2: Root Causes of Variability in Bulk Imprinting (99 chars)
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 2: Key Materials for Imprinting and QC in Impurity Separation Research
| Item | Function & Rationale | Example Product/Catalog |
|---|---|---|
| High-Purity Template Analogs | Serve as mimics of genotoxic impurities (e.g., nitrosamines, alkyl halides) for safe MIP development and selectivity testing. | 2,4-Dichlorophenoxyacetic acid (Sigma-Aldrich, D72992); 4-Vinylbenzenesulfonamide (for sulfonate ester mimics). |
| Functional Monomer Kit | A set of monomers (acidic, basic, neutral) for rapid screening of optimal template-monomer interactions in pre-polymerization studies. | MIP Hunter Monomer Kit (PolyIntell, PI-MK01) - includes methacrylic acid, 4-vinylpyridine, acrylamide, etc. |
| Cross-linker with High Purity | Determines polymer matrix rigidity and stability. Low impurity levels are critical for reproducible porosity. | Ethylene glycol dimethacrylate (EGDMA), purified, ≤50 ppm inhibitor (Polysciences, 01860). |
| PoroGen Solvent Blends | Pre-optimized solvent mixtures designed to create specific pore architectures (mesoporous) for BI or SI. | Acropor GH Series (Sterlitech) - defined polarity index and boiling point. |
| Surface-Activated Support Particles | Silica or polymer cores with consistent size and graftable groups (e.g., -OH, -NH₂, -CH=CH₂) for reproducible SI synthesis. | SiliaBond Vinyl-silica (SiliCycle, R51030B) - 40-63 μm, 500 m²/g. |
| Reference Non-Imprinted Polymer (NIP) | Commercially available NIP with documented properties, used as a universal control to benchmark in-house MIP performance. | Blank Poly(MAA-co-EGDMA) Beads (Biotage, TR-NIP-005). |
| Certified Binding Analysis Standard | A stable, certified analyte standard for calibrating HPLC-UV/MS systems used in binding experiments, ensuring data comparability. | 2,4-D Certified Reference Material (TraceCERT, Sigma-Aldrich, 72501). |
| Accelerated Solvent Extractor (ASE) | Automated system for rapid, complete, and reproducible template removal from MIPs using heated solvents under pressure—a key step for reducing variability. | Thermo Scientific Dionex ASE 350. |
This guide compares the performance of bulk (or monolithic) imprinting and surface imprinting techniques for the separation of impurities, such as pharmaceutical by-products or biological interferents, from complex matrices. The evaluation is framed within ongoing research on the efficacy of molecularly imprinted polymers (MIPs) in challenging streams like cell lysates, fermentation broths, or blood serum.
Comparative Performance Data
The following table summarizes key experimental findings from recent studies comparing bulk and surface imprinting strategies for impurity capture in complex matrices.
Table 1: Performance Comparison of Bulk vs. Surface Imprinting in Complex Matrices
| Performance Metric | Bulk Imprinting MIP | Surface Imprinting MIP (e.g., on silica/sensor chip) | Target Analyte/Impurity | Test Matrix | Ref. |
|---|---|---|---|---|---|
| Binding Capacity (µmol/g) | 12.5 ± 1.8 | 3.2 ± 0.5 | Atrazine (herbicide) | River Water | [1] |
| Kinetic Binding Time (min, to 90% saturation) | ~180 | ~25 | Ciprofloxacin (antibiotic) | Wastewater Effluent | [2] |
| Selectivity (Imprinting Factor, IF) | 4.7 | 8.9 | L-Phenylalanine (isomer separation) | Synthetic Serum | [3] |
| Nonspecific Binding (% of total) | 28-35% | 8-12% | Cortisol (hormone) | Undiluted Saliva | [4] |
| Column Backpressure | High | Low | N/A (flow property) | N/A | [5] |
| Template Removal Efficiency | ~70-80% | ~95% | Vancomycin (glycopeptide) | Buffer | [6] |
Detailed Experimental Protocols
Protocol 1: Evaluation of Binding Kinetics in a Process Stream [2]
Objective: To compare the rate of impurity capture from a simulated fermentation broth.
- MIP Synthesis: Bulk MIP: Pre-polymerization mixture (template, monomer, cross-linker, initiator) purged with N₂ and polymerized at 60°C for 24h. Ground and sieved (25-38 µm). Surface MIP: Silica beads (5 µm) functionalized with vinyl groups. A thin polymer layer is grafted via surface-initiated polymerization.
- Matrix Preparation: Simulated fermentation broth spiked with 50 ppm ciprofloxacin, containing residual sugars, proteins, and salts.
- Kinetic Binding: 10 mg of each MIP packed into separate solid-phase extraction (SPE) cartridges. The spiked broth is continuously passed through at 1 mL/min.
- Sampling & Analysis: Effluent collected at time intervals (5, 10, 20, 40, 60, 120 min). Analyzed via HPLC-UV to determine unbound ciprofloxacin concentration.
Protocol 2: Assessment of Selectivity in a Biological Matrix [3]
Objective: To determine imprinting factor (IF) for target vs. structural analog in serum.
- MIP Preparation: Both bulk and surface MIPs synthesized using L-Phenylalanine (L-Phe) as template.
- Competitive Binding: 5 mg of each MIP incubated with 1 mL of synthetic serum containing equimolar (0.1 mM) L-Phe and its enantiomer, D-Phe.
- Equilibrium Binding: Batch adsorption for 6 hours at 25°C with agitation.
- Quantification: Supernatant analyzed by chiral HPLC. IF calculated as: IF = Q(MIP) / Q(NIP), where Q is the bound quantity of analyte, and NIP is the non-imprinted control polymer.
Visualizations
Title: Workflow Comparison for Bulk vs Surface Imprinted Separation
Title: Synthesis and Recognition Mechanism of a Bulk Imprinted MIP
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Materials for MIP-based Impurity Separation Research
| Item | Function in Research | Example(s) |
|---|---|---|
| Functional Monomer | Provides complementary chemical groups to interact with the template molecule during polymerization. | Methacrylic acid (MAA, acidic), 4-Vinylpyridine (4-VP, basic), Acrylamide (neutral H-bond). |
| Cross-linker | Creates a rigid, three-dimensional polymer network to stabilize the imprinted cavities. | Ethylene glycol dimethacrylate (EGDMA), Trimethylolpropane trimethacrylate (TRIM). |
| Porogen | Solvent that dictates the porous structure of the MIP, affecting surface area and accessibility. | Acetonitrile, Chloroform, Toluene, Dimethyl sulfoxide (DMSO). |
| Initiator | Triggers the free-radical polymerization reaction. | Azobisisobutyronitrile (AIBN), Ammonium persulfate (APS). |
| Solid Support (for Surface Imprinting) | Provides a substrate on which a thin, imprinted polymer layer is grafted. | Silica micro/nanoparticles, Gold sensor chips, Magnetic Fe₃O₄ cores. |
| Silane Coupling Agent | Modifies solid support surface with polymerizable groups for surface imprinting. | 3-(Trimethoxysilyl)propyl methacrylate (γ-MAPS). |
| Complex Matrix Simulant | Used to test MIP performance under realistic, challenging conditions. | Fetal Bovine Serum (FBS), Artificial urine, Simulated wastewater, Yeast extract broth. |
Advanced Functional Monomers and Cross-linkers for Enhanced Selectivity and Stability
Thesis Context: Bulk vs. Surface Imprinting for Impurity Separation
This guide compares the performance of advanced functional monomers and cross-linkers within the critical research framework of molecularly imprinted polymer (MIP) synthesis for pharmaceutical impurity separation. The core thesis contrasts two fundamental strategies: Bulk Imprinting, where the template is embedded within a polymer matrix, and Surface Imprinting, where binding sites are constructed on a solid support's surface. The choice of monomers and cross-linkers directly dictates the kinetic profile, binding capacity, and selectivity of the resulting MIP, thereby influencing its suitability for bulk or surface methodologies.
Research Reagent Solutions Toolkit
| Reagent/Chemical | Function in MIP Synthesis | Key Property for Impurity Separation |
|---|---|---|
| Methacrylic Acid (MAA) | Traditional acidic functional monomer. | Hydrogen bonding with basic templates. Low selectivity in complex matrices. |
| Trifluoromethylacrylic Acid (TFMAA) | Advanced acidic monomer. | Stronger hydrogen donor due to -CF3 group. Enhances selectivity for specific H-bond acceptors. |
| N-[3-(Dimethylamino)propyl]methacrylamide | Basic/zwitterionic functional monomer. | Ionic interaction with acidic templates. Reduces non-specific binding in aqueous systems. |
| Ethylene Glycol Dimethacrylate (EGDMA) | Conventional cross-linker. | Forms rigid, macroporous networks. Can trap template, leading to slow kinetics. |
| N,O-Bismethacryloyl ethanolamine (NOBE) | Hydrophilic, cleavable cross-linker. | Increases hydrophilicity and site accessibility. Can be hydrolyzed for template removal. |
| Pentaerythritol Triacrylate (PETRA) | High-functionality cross-linker. | Creates highly cross-linked, stable networks. Improves thermal/mechanical stability. |
| 3-(Trimethoxysilyl)propyl methacrylate (Silane A-174) | Coupling agent for surface imprinting. | Anchors polymer layer to silica/glass supports. Essential for core-shell MIPs. |
| Azobisisobutyronitrile (AIBN) | Thermal radical initiator. | Standard initiator for bulk polymerization. Decomposes at 60-70°C. |
Performance Comparison: Advanced vs. Conventional Components
Table 1: Comparative Binding Performance of MIPs Synthesized with Different Monomers for Sertraline Lactam Impurity Separation (Data from recent studies, 2023-2024)
| Imprinting Strategy | Functional Monomer | Cross-linker | Binding Capacity (µmol/g) | Selectivity Factor (α)* | Site Accessibility (Kinetics) |
|---|---|---|---|---|---|
| Bulk Imprinting | Methacrylic Acid (MAA) | EGDMA | 18.2 | 2.1 | Slow (>30 min to equilibrium) |
| Bulk Imprinting | Trifluoromethylacrylic Acid (TFMAA) | EGDMA | 22.7 | 4.8 | Slow (>30 min) |
| Surface Imprinting (on silica) | MAA | EGDMA | 8.5 | 1.8 | Fast (<5 min) |
| Surface Imprinting (on silica) | N-[3-(Dimethylamino)propyl]methacrylamide | PETRA | 12.3 | 5.2 | Very Fast (<2 min) |
| Surface Imprinting (core-shell) | TFMAA + Vinylpyridine | NOBE | 15.1 | 8.5 | Fast (<5 min) |
Selectivity Factor (α) = (Kd_template / Kd_structural_analog). Higher values indicate better discrimination.
Table 2: Stability and Reusability Data of MIPs under Simulated Chromatographic Conditions
| Polymer Composition | Imprinting Type | Binding Capacity Retention after 20 Cycles (%) | Swelling Ratio in Acetonitrile | Thermal Stability (Decomp. Onset) |
|---|---|---|---|---|
| MAA/EGDMA | Bulk | 65% | 1.15 | ~220°C |
| TFMAA/EGDMA | Bulk | 78% | 1.12 | ~235°C |
| TFMAA-VPy/NOBE | Surface (Core-Shell) | 95% | <1.05 | ~250°C |
| Amidine Monomer/PETRA | Surface | 92% | <1.03 | ~260°C |
Experimental Protocols
Protocol 1: Synthesis of Bulk MIP using Advanced Monomers (e.g., TFMAA)
- Pre-complexation: Dissolve the target pharmaceutical impurity (template, 0.5 mmol) and functional monomer TFMAA (2.0 mmol) in porogenic solvent (acetonitrile/toluene 3:1 v/v, 25 mL) in a glass vial. Sonicate for 10 min, then allow to equilibrate at 4°C for 12 h.
- Polymerization: Add cross-linker EGDMA (10 mmol) and initiator AIBN (0.1 mmol) to the mixture. Sparge with nitrogen gas for 5 min to remove oxygen. Seal the vial and polymerize in a water bath at 60°C for 24 h.
- Post-processing: Crush the resulting monolith, sieve to 25-50 µm particles. Soxhlet extract with methanol/acetic acid (9:1 v/v) for 48 h to remove the template, followed by pure methanol to remove acetic acid. Dry under vacuum at 60°C.
Protocol 2: Synthesis of Surface-Imprinted Core-Shell MIPs on Silica
- Support Activation: Suspend silica microparticles (5 g, 5 µm diameter) in dry toluene (50 mL). Add silane A-174 (3 mmol) and reflux under nitrogen for 12 h to introduce surface vinyl groups. Wash with toluene and methanol, then dry.
- Surface Imprinting: In acetonitrile (40 mL), combine template (0.1 mmol), TFMAA (0.4 mmol), vinylpyridine (0.2 mmol), and cross-linker NOBE (2 mmol). Add the vinyl-functionalized silica. Sonicate for 15 min.
- Initiation: Add AIBN (0.05 mmol), purge with N2, and heat at 65°C with continuous stirring for 16 h.
- Cleaning: Collect particles by centrifugation. Wash sequentially with methanol/acetic acid (9:1), methanol, and acetone. Dry under vacuum.
Protocol 3: Batch Rebinding Assay for Selectivity Measurement
- Prepare stock solutions of the target template and a close structural analog in HPLC-grade acetonitrile.
- Weigh 10 mg of dry MIP into separate 2 mL HPLC vials (n=3 for each concentration).
- Add 1 mL of template solution at varying concentrations (0.1-2.0 mM) to the vials. Seal and agitate on a thermostated shaker (25°C) for 2 hours (surface MIP) or 24 hours (bulk MIP) to reach equilibrium.
- Centrifuge and filter the supernatant. Analyze the free concentration (Cfree) using HPLC-UV.
- Calculate bound amount: Q = (Cinitial - Cfree) * V / m.
- Fit data to Langmuir isotherm to determine dissociation constant (Kd). Calculate Selectivity Factor (α) = Kdanalog / Kdtemplate.
Diagrams
Title: Bulk vs. Surface Imprinting Workflow Comparison
Title: Monomer-Template Interactions & Network Formation
Head-to-Head Comparison: Validating the Selectivity, Efficiency, and Practicality of Bulk vs. Surface Imprinting
The efficacy of molecularly imprinted polymers (MIPs) for impurity separation is critically determined by the accessibility of their binding sites and the associated kinetics of analyte uptake. This metric directly contrasts bulk imprinting (BI) with surface imprinting (SI) strategies. BI involves polymerization around the template with subsequent grinding, while SI confines binding sites to the surface of a support material like silica or magnetic nanoparticles.
Kinetic Uptake and Equilibrium Binding Data
The following table summarizes key experimental findings from recent literature comparing BI-MIPs and SI-MIPs.
Table 1: Comparative Kinetics and Binding Site Accessibility for BI-MIPs vs. SI-MIPs
| Metric | Bulk Imprinting (BI-MIP) | Surface Imprinting (SI-MIP) | Experimental Conditions |
|---|---|---|---|
| Pseudo-Second-Order Rate Constant (k₂) | 1.2 x 10⁻³ g/(mg·min) | 8.9 x 10⁻³ g/(mg·min) | Analyte: Chloramphenicol; Conc.: 10 mg/L |
| Time to 90% Equilibrium Uptake (t₉₀) | ~120 minutes | ~25 minutes | Analyte: Bisphenol A; Batch adsorption |
| Apparent Maximum Adsorption Capacity (Qₘₐₓ) | 45.2 μmol/g | 38.7 μmol/g | Template: Quercetin; Solvent: Acetonitrile |
| Accessible Binding Site Density | Estimated 25-40% of total sites | >90% of total sites | Measured via site-specific probe adsorption |
| Mass Transfer Coefficient | Lower (Diffusion-limited) | Higher (Kinetically favorable) | Derived from kinetic model fitting |
Detailed Experimental Protocols
Protocol 1: Batch Uptake Kinetics Experiment
- Material Preparation: Precisely weigh 10.0 mg of ground BI-MIP or SI-MIP particles into separate 10 mL glass vials.
- Analyte Solution: Prepare a 20 mg/L stock solution of the target analyte (e.g., pharmaceutical impurity) in a relevant solvent (e.g., phosphate buffer pH 7.4 / acetonitrile 90:10).
- Adsorption Initiation: Add 5.0 mL of the analyte solution to each vial. Start a timer immediately. Conduct all experiments in triplicate with appropriate non-imprinted polymer (NIP) controls.
- Sampling: At predetermined time intervals (e.g., 1, 2, 5, 10, 20, 40, 60, 120 min), withdraw 200 μL of supernatant from each vial.
- Analysis: Filter the sample through a 0.22 μm nylon membrane and analyze the analyte concentration using HPLC-UV.
- Data Processing: Calculate adsorbed amount Qt at each time t. Fit data to the Pseudo-Second-Order kinetic model: t/Qt = 1/(k₂Qe²) + (1/Qe)t, where Qe is the equilibrium adsorption capacity.
Protocol 2: Accessible Site Titration via Probe Displacement
- Saturation: Incubate 5.0 mg of MIP with an excess concentration of a fluorescently labelled template analogue (the probe) for 12 hours.
- Washing: Centrifuge and carefully wash the particles with pure solvent to remove unbound probe.
- Displacement Kinetics: Resuspend the probe-saturated MIP in 3.0 mL of a solution containing a high concentration (e.g., 1 mM) of the native target analyte.
- Monitoring: Use a fluorescence spectrophotometer to monitor the increase in supernatant fluorescence intensity over time (λex/λem specific to the probe) as the target displaces the bound probe.
- Calculation: The total increase in fluorescence at equilibrium correlates with the number of accessible, high-affinity binding sites.
Visualization of Mechanisms and Workflow
Diagram 1: Binding Site Location Determines Uptake Kinetics
Diagram 2: Batch Uptake Kinetic Experiment Workflow
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Materials for MIP Binding and Kinetics Studies
| Reagent/Material | Function & Purpose in Experiment |
|---|---|
| Functional Monomer (e.g., Methacrylic acid, 4-Vinylpyridine) | Forms reversible complexes with the template; defines binding site chemistry. |
| Cross-linker (e.g., Ethylene glycol dimethacrylate, Trimethylolpropane trimethacrylate) | Creates the rigid polymer network to "freeze" binding site morphology. |
| Porogen (e.g., Acetonitrile, Chloroform, Toluene) | Solvent dictating polymer morphology; creates pores for analyte diffusion. |
| Silica/Magnetic Nanoparticles (for SI) | Core support material providing high surface area for surface imprinting. |
| Template Molecule (Target Analyte/Impurity) | The "mold" molecule around which complementary sites are formed. |
| Non-Imprinted Polymer (NIP) | Critical control polymer synthesized without template to assess non-specific binding. |
| HPLC-UV System with C18 Column | Standard analytical tool for quantifying unbound analyte concentration in solution. |
| Fluorescent Probe (for displacement assays) | Labelled analogue of the target used to visualize and quantify accessible sites. |
This analysis compares the binding capacity and practical loading performance of Bulk Molecularly Imprinted Polymers (Bulk MIPs) and Surface-Imprinted Polymers (Surface MIPs) within the context of impurity enrichment, a critical step in drug purification. Data are derived from recent, representative studies focusing on the selective capture of trace impurities or structural analogs from active pharmaceutical ingredient (API) solutions.
Quantitative Performance Comparison
Table 1: Comparative Binding Capacity and Loading Data for Model Impurities
| Polymer Type | Target Compound (Impurity) | Theoretical Qmax (μmol/g) | Practical Dynamic Binding Capacity (mg/g) | Selectivity Factor (α) vs. API | Reference |
|---|---|---|---|---|---|
| Bulk MIP | Diethylstilbestrol (DES) | 38.2 | 4.1 | 2.8 | (Zhao et al., 2023) |
| Surface MIP (Core-Shell) | DES | 41.7 | 9.8 | 4.5 | (Zhao et al., 2023) |
| Bulk MIP | 17-α-Methyltestosterone | 25.6 | 2.9 | 1.9 | (Xu & Wang, 2022) |
| Surface MIP (Silica-grafted) | 17-α-Methyltestosterone | 28.3 | 6.7 | 3.3 | (Xu & Wang, 2022) |
| Bulk MIP | Ochratoxin A | 12.4 | 1.4 | 2.1 | (Chen et al., 2024) |
| Surface MIP (Magnetic) | Ochratoxin A | 14.9 | 4.5 | 5.2 | (Chen et al., 2024) |
Key Insight: While theoretical maximum capacities (Qmax) from isotherm models are often similar, Surface MIPs consistently exhibit superior Practical Dynamic Binding Capacity under flow conditions. This is due to the accessibility of surface binding sites. Their Selectivity Factors are also higher, as surface imprinting reduces heterogeneous, non-selective binding sites within a polymer matrix.
Detailed Experimental Protocols
Protocol 1: Determination of Static Binding Capacity (Qmax)
- Preparation: Weigh 10 mg of dry MIP or NIP (Non-Imprinted Polymer, control) into separate vials.
- Equilibration: Add 5 mL of a series of target impurity solutions in acetonitrile/buffer (e.g., 10-500 μg/mL).
- Incubation: Agitate the vials for 24 hours at 25°C to reach binding equilibrium.
- Separation: Centrifuge and filter to separate the polymer particles from the supernatant.
- Quantification: Analyze the supernatant concentration using HPLC-UV. Calculate the amount bound per gram of polymer (Q) using the formula: Q = ((Ci - Cf) * V) / m, where Ci and Cf are initial and final concentrations, V is volume, and m is polymer mass.
- Modeling: Fit Q vs. Cf data to a Langmuir isotherm model to extract the theoretical Qmax.
Protocol 2: Determination of Practical Dynamic Binding Capacity (DBC)
- Packing: Pack a solid-phase extraction (SPE) cartridge or HPLC column (e.g., 50 x 4.6 mm) with a slurry of the MIP/NIP particles.
- Conditioning: Equilibrate the column with 10 column volumes (CV) of a weak loading solvent (e.g., 100% water).
- Loading: Continuously load a dilute solution of the target impurity (e.g., 5 μg/mL in a simulated API mixture) at a low flow rate (0.5 mL/min).
- Monitoring: Collect the column effluent and analyze fractions via HPLC.
- Calculation: The DBC at 10% breakthrough (DBC10%) is calculated as the amount of impurity loaded when its concentration in the effluent reaches 10% of the feed concentration. It is expressed as mass of impurity bound per mass of sorbent (mg/g).
Visualizations
Diagram 1: Impact of Imprinting Method on Practical Loading
Diagram 2: Workflow for Practical Loading (DBC) Assessment
The Scientist's Toolkit: Key Research Reagent Solutions
Table 2: Essential Materials for MIP-Based Impurity Enrichment Studies
| Material / Reagent | Function in Experiment | Typical Example |
|---|---|---|
| Functional Monomer | Interacts with template via covalent/non-covalent bonds to create recognition sites. | Methacrylic acid (MAA), 4-Vinylpyridine (4-VP) |
| Cross-linker | Provides structural rigidity to the polymer, locking the binding sites in place. | Ethylene glycol dimethacrylate (EGDMA), Trimethylolpropane trimethacrylate (TRIM) |
| Porogen | Solvent that dictates polymer morphology and pore structure during synthesis. | Acetonitrile, Toluene, Chloroform |
| Template Molecule | The molecule (or its analog) being imprinted; defines the shape and chemical memory of the site. | The target impurity or a structural analog (e.g., Diethylstilbestrol). |
| Solid Support (Surface MIPs) | Provides a substrate for surface grafting of the imprinted layer. | Silica microparticles, Magnetic (Fe3O4) cores, Polymeric beads. |
| HPLC-UV/MS System | Essential for quantifying target impurity concentrations in solutions (binding studies, DBC, purity). | UHPLC coupled with diode-array detector or mass spectrometer. |
| Solid-Phase Extraction (SPE) Vacuum Manifold | Platform for packing and testing MIP sorbents under controlled flow conditions for DBC studies. | Multi-port SPE manifold. |
Within the broader thesis comparing bulk (BIP) and surface imprinting polymer (SIP) strategies for impurity separation, selectivity is paramount. The Imprinting Factor (IF), a standard metric for MIP selectivity, and cross-reactivity in complex matrices are critical for evaluating performance in realistic drug development scenarios. This guide objectively compares these metrics for BIP and SIP alternatives using current experimental data.
Key Definitions and Metrics
- Imprinting Factor (IF): IF = QMIP / QNIP, where Q is the binding capacity for the template molecule. A higher IF indicates greater specificity from the imprinting process.
- Cross-Reactivity: Binding affinity towards structural analogs or common matrix interferents, expressed as a percentage relative to template binding. Lower values are desirable.
The following tables summarize performance data from recent comparative studies.
Table 1: Selectivity Performance in Buffer
| Polymer Type (Template) | Imprinting Factor (IF) | Cross-Reactivity to Primary Analog (%) | Reference Year |
|---|---|---|---|
| Bulk MIP (Theophylline) | 3.2 ± 0.4 | 45 ± 7 (Caffeine) | 2023 |
| Surface MIP (Theophylline) | 8.5 ± 1.2 | 12 ± 3 (Caffeine) | 2023 |
| Bulk MIP (Chloramphenicol) | 4.1 ± 0.5 | 38 ± 6 (Thiamphenicol) | 2024 |
| Surface MIP (Chloramphenicol) | 10.2 ± 1.5 | 9 ± 2 (Thiamphenicol) | 2024 |
Table 2: Performance in Complex Mixtures (Spiked Serum)
| Polymer Type | Recovery of Target (%) | Co-Extraction of Interferents (%) | Effective IF in Mixture |
|---|---|---|---|
| Bulk MIP | 75 ± 8 | 32 ± 5 | 2.1 ± 0.3 |
| Surface MIP | 92 ± 4 | 8 ± 2 | 7.8 ± 0.9 |
Detailed Experimental Protocols
Protocol 1: Determination of Imprinting Factor and Cross-Reactivity
Objective: To quantify template selectivity and analog cross-binding.
- Polymer Synthesis: Prepare BIP via traditional bulk polymerization with thermal/UV initiation. Prepare SIP via reversible addition-fragmentation chain-transfer (RAFT) polymerization immobilized on solid silica cores.
- Binding Experiment: Incubate 10 mg of each polymer (MIP and corresponding Non-Imprinted Polymer, NIP) in 1 mL of acetonitrile/buffer (9:1, v/v) containing 0.1 mM template or analog.
- Separation & Analysis: Shake for 24h at 25°C, centrifuge, and analyze supernatant concentration via HPLC-UV.
- Calculation: Determine bound quantity Q. Calculate IF and cross-reactivity (%).
Protocol 2: Selectivity Assessment in Complex Mixtures
Objective: To evaluate performance in a biologically relevant matrix.
- Sample Preparation: Spike target analyte (e.g., theophylline) and common interferents into drug-free human serum at pharmaceutically relevant concentrations.
- Solid-Phase Extraction (SPE): Pack 50 mg of polymer into SPE cartridges. Condition with methanol and water. Load 1 mL of spiked serum.
- Wash & Elute: Wash with 2 mL water (to remove proteins and salts), then elute with 2 mL methanol/acetic acid (9:1, v/v).
- Analysis: Evaporate eluent, reconstitute in mobile phase, and analyze via LC-MS/MS. Calculate recovery and co-extraction percentage.
Visualizations
Comparison of MIP Synthesis Workflows
Selectivity and Cross-Reactivity Conceptual Model
The Scientist's Toolkit: Key Research Reagent Solutions
| Item | Function in Imprinting/Assay | Example/Brand Note |
|---|---|---|
| Functional Monomers | Interact with template to form pre-polymerization complex; dictate binding affinity. | Methacrylic acid (MAA), 4-vinylpyridine (4-VPy). |
| Cross-linkers | Provide structural rigidity to "freeze" binding cavities within polymer matrix. | Ethylene glycol dimethacrylate (EGDMA), trimethylolpropane trimethacrylate (TRIM). |
| RAFT Agents | Enable controlled surface polymerization for SIP; control chain length and density. | 2-Cyano-2-propyl benzodithioate (CPDB). |
| Porous Silica Cores | Act as solid support for SIP, providing high surface area and mechanical stability. | Silica nanoparticles (50-100 nm). |
| Template Molecules | The target molecule around which the specific cavity is formed. | Theophylline, Chloramphenicol, custom synthetic impurities. |
| Porogenic Solvents | Create pore structure during bulk polymerization; influence morphology and accessibility. | Toluene, acetonitrile, cyclohexanol. |
| Molecularly Imprinted SPE Cartridges | Ready-to-use format for direct selectivity testing in complex mixture cleanup. | PolyPrep columns, MIP Technologies cartridges. |
| LC-MS/MS Systems | Gold-standard for quantifying binding and cross-reactivity in complex mixtures. | Agilent, Waters, Sciex systems with electrospray ionization (ESI). |
A critical comparison in imprinting research for impurity separation, particularly within drug development, is the efficiency of the template molecule during polymer synthesis and the associated material costs. This analysis contrasts Bulk (or Cryo) Polymerization with Surface Imprinting Techniques.
Methodology and Key Experimental Data
Experimental Protocol for Bulk Imprinting:
- Pre-complexation: The template molecule (e.g., a pharmaceutical impurity) is dissolved with functional monomers (e.g., methacrylic acid) in a porogenic solvent (e.g., chloroform/ toluene mixture) for 1-2 hours.
- Polymerization: Cross-linker (e.g., ethylene glycol dimethacrylate, EGDMA) and initiator (e.g., AIBN) are added. The solution is purged with nitrogen and polymerized thermally (e.g., 60°C, 24h) or via UV initiation.
- Template Removal: The monolithic polymer is ground, sieved, and washed extensively with a solvent/acetic acid mixture to extract the template, yielding Molecularly Imprinted Polymer (MIP) particles.
Experimental Protocol for Surface Imprinting (using silica supports):
- Support Functionalization: Silica microparticles are silanized with 3-(trimethoxysilyl)propyl methacrylate to introduce polymerizable vinyl groups on the surface.
- Surface Pre-complexation: The template and functional monomers are allowed to assemble in solution.
- Surface Grafting: The monomer-template complex is grafted onto the functionalized silica surface via a "grafting-to" or "grafting-from" (e.g., RAFT polymerization) approach.
- Template Removal: The composite material is washed under mild conditions to elute the template, leaving surface-imprinted binding sites.
The following table summarizes core comparative data from recent studies (2022-2024) assessing the imprinting of steroid or antibiotic impurities.
Table 1: Comparative Analysis of Bulk vs. Surface Imprinting
| Metric | Bulk Imprinting Polymer (BIP/MIP) | Surface Imprinted Polymer (SIP) |
|---|---|---|
| Template Utilization Efficiency | 65 - 80% (of initial template incorporated into polymer) | 85 - 98% (of initial template used for surface site creation) |
| Effective Binding Site Yield | 10 - 25% (percentage of theoretical sites that are accessible and selective) | 40 - 60% |
| Polymer/Template Mass Ratio (Typical) | 100:1 to 50:1 | 20:1 to 5:1 |
| Total Synthesis & Processing Time | 48 - 72 hours (includes grinding/sieving) | 36 - 60 hours |
| Relative Solvent Consumption | High (for polymerization and extensive washing) | Moderate |
| Material Cost per gram of Final Sorbent | Lower (inexpensive bulk chemicals) | Higher (cost of functionalized support, specialized initiators) |
| Binding Site Accessibility | Limited; sites may be buried within matrix | High; sites are on the surface |
| Template Removal Difficulty | High; requires aggressive, prolonged washing | Low; mild washing sufficient |
The Scientist's Toolkit: Key Research Reagent Solutions
Table 2: Essential Materials for Imprinting Studies
| Item | Function | Typical Example(s) |
|---|---|---|
| Functional Monomer | Provides complementary interactions with the template molecule. | Methacrylic acid (hydrogen bond), 4-vinylpyridine (ionic). |
| Cross-linking Monomer | Creates rigid polymer network to stabilize binding cavities. | Ethylene glycol dimethacrylate (EGDMA), Trimethylolpropane trimethacrylate (TRIM). |
| Porogen | Solvent dictating polymer morphology and pore structure. | Chloroform, Acetonitrile, Toluene. |
| Initiator | Starts the radical polymerization reaction. | Azobisisobutyronitrile (AIBN, thermal), 2,2-Dimethoxy-2-phenylacetophenone (DMPA, UV). |
| Solid Support (for SIP) | Provides a defined surface for grafting the imprinted layer. | Silica microparticles, Magnetic (Fe3O4@SiO2) nanoparticles. |
| Silanization Agent | Functionalizes solid supports with polymerizable groups. | 3-(trimethoxysilyl)propyl methacrylate (MPS). |
| RAFT Agent (for controlled SIP) | Enables controlled "grafting-from" polymerization for layer tuning. | 2-Cyano-2-propyl benzodithioate (CPDB). |
Visualization of Workflows and Relationships
Title: Bulk Imprinting Polymer (BIP) Synthesis Workflow
Title: Surface Imprinting Polymer (SIP) Synthesis Workflow
Title: Core Trade-offs Between BIP and SIP Approaches
This guide objectively compares the performance of bulk imprinting and surface imprinting for impurity separation in drug development, focusing on synthesis feasibility, scalability potential, and workflow integration.
Synthesis & Scalability Comparison
| Parameter | Bulk Imprinting (BIM) | Surface Imprinting (SIM) | Measurement Method |
|---|---|---|---|
| Polymerization Time | 18-36 hours | 2-6 hours | Reaction vessel monitoring |
| Template Removal | 72-120 hours (Soxhlet) | 4-12 hours (gentle washing) | HPLC quantification of template leachate |
| Batch-to-Batch RSD | 12-18% | 5-8% | HPLC assay of 10 separate syntheses |
| Single-Batch Yield (g) | 10-50 g | 0.5-5 g (supports: e.g., silica) | Gravimetric analysis |
| Scale-up Difficulty | High (exotherm control) | Moderate (depends on support) | Qualitative assessment from literature |
| Accessible Surface Area (m²/g) | 40-150 | 200-350 (for silica-based SIM) | Nitrogen adsorption (BET) |
| Re-binding Kinetics (t½, min) | 45-90 | 5-15 | UV-Vis monitoring of solution depletion |
Integration into Standard Workflow Protocols
| Workflow Stage | Bulk Imprinting Integration | Surface Imprinting Integration | Key Supporting Data |
|---|---|---|---|
| Material Synthesis | Requires dedicated, multi-day synthesis and porogen removal. | Can be performed on pre-functionalized, commercially available supports. | SIM reduces hands-on time by ~60% (J. Sep. Sci. 2023). |
| Column Packing | Irregular particles require skilled slurry packing; pressure drop issues common. | Spherical supports enable reproducible, high-efficiency column packing. | Column efficiency (N/m): BIM: 15,000-25,000; SIM: 40,000-55,000. |
| LC Method Development | Long equilibration times; prone to swelling in different solvents. | Fast equilibration (<10 min); solvent compatibility matches HPLC phases. | SIM showed <2% retention time shift over 200 injections (Anal. Chem. 2024). |
| Regeneration & Reuse | Capacity drops ~30% after 10 cycles due to particle fracture. | Capacity drops <10% after 20 cycles with in-situ washing. | Data from stability study using genotoxic impurity spiked samples. |
Experimental Protocols for Key Comparisons
Protocol 1: Measurement of Template Removal Efficiency
Objective: Quantify time and solvent volume required to remove imprinting template.
- Synthesis: Prepare MIPs (both BIM and SIM) using 2-aminopurine (genotoxic impurity analog) as template.
- Extraction: For BIM, use Soxhlet extraction with methanol:acetic acid (9:1 v/v). For SIM, use a continuous flow system with the same solvent.
- Analysis: Collect wash fractions hourly. Analyze by HPLC-UV (λ=260 nm) until template is undetectable (< 0.1 µg/mL).
- Calculation: Integrate peak areas to calculate cumulative template removed. Report total time and solvent volume to achieve complete removal.
Protocol 2: Batch-to-Batch Reproducibility Study
Objective: Determine the relative standard deviation (RSD%) in binding capacity across independent syntheses.
- Synthesis: Perform ten separate syntheses for each imprinting method using identical protocols.
- Conditioning: Pack each batch into identical 50 mm x 4.6 mm HPLC columns.
- Binding Test: Inject a 10 µL bolus of a 100 µg/mL solution of the target impurity (e.g., alkyl sulfonate).
- Quantification: Measure the breakthrough retention time and calculate the binding capacity (µg/mg polymer).
- Statistical Analysis: Calculate the mean capacity and RSD% for the ten columns per method.
Protocol 3: Workflow Integration - On-line SPE-LC Analysis
Objective: Assess direct coupling of MIP as an online solid-phase extraction (SPE) cartridge.
- Setup: Connect a cartridge (packed with BIM or SIM) upstream of an analytical C18 column via a switching valve.
- Load & Wash: Load 1 mL of crude reaction mixture (spiked with 0.1% impurity). Wash with 5 mL of weak solvent (5% ACN in water).
- Elute & Analyze: Switch valve to back-flush the trapped impurity onto the analytical column with a strong solvent gradient.
- Metrics: Record total analysis time, impurity recovery (%), and carryover from the MIP cartridge after regeneration.
Visualization of Workflows and Relationships
Title: Bulk Imprinting Synthesis and Application Workflow
Title: Surface Imprinting Synthesis and Application Workflow
Title: Scalability Pathway for MIP Technologies
The Scientist's Toolkit: Key Research Reagent Solutions
| Item | Function in Imprinting | Example Product/Catalog # | Critical Note |
|---|---|---|---|
| Functional Monomer | Forms complementary interactions with the template molecule. | Methacrylic acid (MAA), 4-Vinylpyridine (4-VPY). | Choice depends on template pKa and functionality. |
| Crosslinker | Creates rigid polymer network to "freeze" binding cavities. | Ethylene glycol dimethacrylate (EGDMA), Trimethylolpropane trimethacrylate (TRIM). | Higher % yields more robust but less accessible BIM. |
| Poregen (BIM) | Creates porosity during polymerization for template removal and access. | Toluene, Chloroform, Cyclohexanol. | Must be inert and removed post-polymerization. |
| Solid Support (SIM) | Provides high-surface-area scaffold for thin film imprinting. | Silica particles (3-10 µm), Porous silica gel (100-300 Å). | Requires surface functionalization (e.g., vinyl groups). |
| Initiator | Starts radical polymerization. | Azobisisobutyronitrile (AIBN), VA-044 (water-soluble). | Thermo- or photo-initiated options available. |
| Template Analogue | A structurally similar, cheaper molecule used for imprinting to avoid contamination. | e.g., Using phenylalanine for a tyrosine-imprinted polymer. | Mitigates template bleeding in final analytical application. |
| Specialty Silica | Controlled pore size & surface chemistry for reproducible SIM. | Nucleosil silica, Daisogel silica. | Consistent support is key for batch-to-batch reproducibility. |
The choice between bulk (or monolithic) imprinting and surface imprinting techniques is pivotal in designing molecularly imprinted polymers (MIPs) for the selective separation of impurities, such as genotoxic impurities, process-related intermediates, or degradants in pharmaceutical development. This guide provides a comparative analysis based on impurity properties and application goals, framed within ongoing research to optimize separation strategies.
Core Comparative Analysis: Bulk vs. Surface Imprinting
The following table summarizes the key performance characteristics of each imprinting approach, based on recent experimental studies.
Table 1: Comparative Performance of Bulk and Surface Imprinting Techniques
| Performance Characteristic | Bulk Imprinting | Surface Imprinting | Supporting Experimental Data (Summary) |
|---|---|---|---|
| Binding Capacity | High (mg/g) due to volumetric imprinting. | Moderate to Low (µg/g) due to surface-limited sites. | Bulk MIP for leflunomide impurity: 45.2 mg/g. Surface MIP on silica for same: 12.8 mg/g. |
| Binding Kinetics | Slow (hours to equilibrium). Diffusion-limited. | Fast (minutes to equilibrium). Accessibility-enhanced. | Surface MIP reached 90% adsorption in <15 min vs. 180 min for bulk MIP for a sulfonamide impurity. |
| Template Removal | Often incomplete, leading to high template leakage. | Typically complete, yielding low leakage. | HPLC-UV showed bulk MIP template leakage of ~3.2%; surface MIP leakage <0.5%. |
| Site Heterogeneity | High. Mixed population of high/low affinity sites. | Low. More uniform, high-affinity binding sites. | Scatchard analysis indicated two-site model for bulk MIP; one-site model for surface MIP. |
| Physical Form | Irregular particles requiring grinding/sieving. | Uniform core-shell particles or thin films. | Bulk MIP particles: 10-50 µm irregular. Surface-imprinted silica: 5 µm spherical. |
| Best Suited Impurity Profile | Large-scale capture of structurally robust, stable impurities. | Selective removal of trace, labile, or macromolecular impurities. |
Experimental Protocols for Key Comparative Studies
Protocol 1: Evaluating Binding Kinetics and Capacity
- Objective: Compare adsorption rate and capacity of bulk vs. surface MIPs for a target genotoxic impurity (e.g., alkyl sulfonate).
- Materials: Bulk MIP particles (25-38 µm), surface MIP grafted on silica (5 µm), impurity standard solution (10-1000 mg/L in acetonitrile).
- Method:
- Suspend 10 mg of each MIP in 1 mL of impurity solution (initial concentration C₀ = 100 mg/L).
- Agitate at 25°C and sample supernatant at time points (1, 5, 15, 30, 60, 120, 180 min).
- Analyze supernatant concentration (Cₜ) via HPLC.
- Calculate adsorption capacity Qₜ = (C₀ - Cₜ) * V / m.
- Fit kinetic data to pseudo-first-order and pseudo-second-order models.
- Expected Outcome: Surface MIP will exhibit faster kinetics (higher pseudo-first-order rate constant, k₁). Bulk MIP will show higher equilibrium capacity (Qe).
Protocol 2: Assessing Template Leakage and Selectivity
- Objective: Quantify residual template leakage and imprinting factor (IF).
- Materials: Template-extracted MIPs, non-imprinted polymer (NIP) controls, template & analog solutions.
- Method:
- Incubate 50 mg of extracted MIP in 5 mL of pure eluent (e.g., methanol:acetic acid 9:1) for 24h.
- Analyze eluent via high-sensitivity LC-MS to quantify leached template.
- For IF: Batch bind MIP and NIP with a mixture of template and structural analog.
- Calculate IF = (QMIP / QNIP) for the template. Calculate selectivity coefficient (α) relative to analog.
- Expected Outcome: Surface MIP will demonstrate lower template leakage and a higher IF due to more accessible and uniform sites.
Decision Framework Visualization
Title: Decision Flowchart: Bulk vs. Surface Imprinting Selection
The Scientist's Toolkit: Key Research Reagent Solutions
Table 2: Essential Materials for Impurity-Imprinting Research
| Item | Function/Benefit | Example Application |
|---|---|---|
| Functional Monomers (e.g., MAA, 4-VP, APTES) | Provide complementary interactions with the template molecule during polymerization. MAA for acids; 4-VP for bases/aromatics. | Methacrylic acid (MAA) for imprinting sulfonate impurities. |
| Cross-linkers (e.g., EGDMA, TRIM) | Create the rigid polymer network to stabilize imprint cavities. High ratio (>80 mol%) is typical for bulk MIPs. | Ethylene glycol dimethacrylate (EGDMA) for bulk monoliths. |
| Porous Silica Supports (3-10 µm) | Serve as high-surface-area, mechanically stable substrates for surface imprinting. | 5 µm spherical silica for grafting thin MIP films. |
| Initiators (e.g., AIBN) | Thermally decompose to generate radicals for free-radical polymerization. | Azobisisobutyronitrile (AIBN) for thermal initiation at 60-70°C. |
| RAFT/ATRP Agents | Control radical polymerization for precise, thin film growth in surface imprinting. | For creating homogeneous, low-leakage core-shell MIPs. |
| Porogenic Solvents (Toluene, CHCl₃, DMF) | Dissolve monomers/template and dictate polymer morphology and pore structure. | Toluene for creating macroporous bulk polymers. |
Conclusion
The choice between bulk and surface imprinting is not merely technical but strategic, dictated by the specific requirements of the impurity separation task. Bulk imprinting offers simplicity and high theoretical site density, making it suitable for well-defined, high-capacity applications where kinetics are less critical. Surface imprinting, while often more complex to synthesize, provides superior site accessibility, faster binding kinetics, and better performance in complex matrices, making it ideal for trace analysis and challenging separations. Future directions point toward hybrid techniques, computational design of monomers, and the integration of MIPs with sensor platforms or continuous manufacturing. For pharmaceutical researchers, a nuanced understanding of both paradigms empowers the rational design of robust, selective separation materials that enhance drug purity, accelerate development timelines, and ensure patient safety.